Abstract
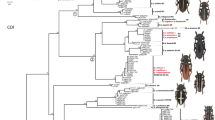
We obtained full (647 bp) or mini (291 bp) DNA barcodes of 140 mostly African and European specimens of 25 Accipiter (Aves: Accipitridae) species. Kimura two-parameter (K2P) distances were calculated between barcodes to determine the thresholds of intra- and interspecific species boundaries. Thresholds were comparable to or higher than those in previous studies and ranged from 2.8 to 3.0 % (best compromise threshold based on cumulative intra- and interspecific K2P distances) and from 3.9 to 5.3 % (ten times the average intraspecific K2P distance). Identification success was determined using the best match and best close-match criteria and ranged between 84 % (mini barcodes) and 90 % (full barcodes). Incorrectly or ambiguously identified specimens belonged to two species that were represented by single sequences in the database (A. madagascariensis and A. trivirgatus) and three species pairs that shared at least one haplotype: viz. A. nisus and A. rufiventris, A. gularis and A. virgatus, and A. cooperii and A. gundlachi. The other 19 species were unambiguously identified using the full DNA barcodes. The studied species belong to eight traditional superspecies, of which three ([gentilis], [cooperii], and [tachiro]) were well supported. In one superspecies, [badius], species pairs were supported but not the superspecies.
Zusammenfassung
DNA-Barcoding und evolutionäre Beziehungen innerhalb der Gattung Accipiter Brisson, 1760 (Aves, Falconiformes: Accipitridae), mit besonderem Schwerpunkt auf deren afrikanischen und eurasischen Vertretern
Wir verwendeten vollständige DNA-Barcodes (647 bp) oder Mini-Barcodes (291 bp) von 140 Individuen (hauptsächlich afrikanischer und europäischer Herkunft) aus 25 Accipiter-Arten (Aves: Accipitridae). Um die Schwellenwerte für die intra- und interspezifischen Artgrenzen zu ermitteln, berechneten wir die Kimura-2-Parameter-Distanzen (K2P) zwischen den Barcodes. Die Schwellenwerte waren vergleichbar oder höher als die aus früheren Studien und lagen zwischen 2,8 und 3,0 % (BCTh-Schwellenwert (Best Compromise Threshold) auf der Grundlage kumulierter intra- und interspezifischer K2P-Distanzen) beziehungsweise zwischen 3,9 und 5,3 % (zehnfacher Durchschnitt der intraspezifischen K2P-Distanz). Der Erfolg der Zuordnung wurde anhand von Best-Match- und Best-Close-Match-Kriterien bestimmt und reichte von 84 % (Mini-Barcodes) bis 90 % (vollständige Barcodes). Nicht korrekt oder nicht eindeutig bestimmte Individuen stammten von zwei Arten, die in der Datenbank jeweils nur durch einzelne Sequenzen vertreten waren, sowie von drei Artenpaaren, welche mindestens einen gemeinsamen Haplotyp aufwiesen, nämlich: A. nisus − A. rufiventris, A. gularis − A. virgatus und A. cooperii − A. gundlachi. Die übrigen 19 Arten konnten anhand der vollständigen DNA-Barcodes eindeutig zugeordnet werden. Die untersuchten Arten gehören zu acht traditionell gebräuchlichen Superspezies, von denen drei ([gentilis], [cooperii] und [tachiro]) gut bestätigt wurden. Für eine Superspezies ([badius]), konnten zwar Artenpaare bestätigt werden, die Superspezies jedoch nicht.





Similar content being viewed by others
References
Aliabadian M, Kaboli M, Nijman V, Vences M (2009) Molecular identification of birds: performance of distance-based DNA barcoding in three genes to delimit parapatric species. PLoS ONE 4:e4119
Ananian V, Aghababyan K, Tumanyan S, Janoyan G, Bildstein K (2010) Shikra Accipiter badius breeding in Armenia. Sandgrouse 32:151–155
Bildstein KL (2004) Raptor migration in the Neotropics: patterns, processes, and consequences. Ornithol Neotropical 15:83–99
Cai Y, Yue H, Jiang W, Xie S, Li J, Zhou S (2010) DNA barcoding on subsets of three families in Aves. mtDNA 21:132–137
Chelomina GN (2006) Ancient DNA. Genetika 42:293–309
DeSalle R, Egan MG, Siddall M (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Phil Trans R Soc B Biol Sci 1462:1905–1916
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ferguson-Lees J, Christie DA (2001) Raptors of the world. Christopher Helm, London
Griffiths CS, Barrowclough GF, Groth JG, Mertz LA (2007) Phylogeny, diversification and classification of the Accipitridae based on DNA sequences of the RAG-1 exon. J Avian Biol 38:587–602
Hajibabei M, Smith MA, Janzen DH, Rodriguez JJ, Whirfield JB, Hebert PDN (2006) A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes 6:959–964
Hawksworth DL (2010) Terms used in bionomenclature. Including terms used in botanical, cultivated plant, phylogenetic, phytosociological, prokaryote (bacteriological), virus, and zoological nomenclature. Global Biodiversity Information Facility, Copenhagen
Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321
Hebert PDN, Stoeckle MY, Zemlak ST, Francis CM (2004) Identification of birds through DNA barcodes. PLoS Biol 2:1657–1663
Hillis DM, Huelsenbeck JP (1992) Signal, noise, and reliability in molecular phylogenetic analyses. J Hered 83:189–195
Johnsen A, Rindal E, Ericson PGP, Zuccon D, Kerr KCR, Stoeckle MY, Lifjeld JT (2010) DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. J Ornith 151:565–578
Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN (2007) Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes 7:535–543
Kerr KCR, Lijtmaer DA, Barreira AS, Hebert PDN, Tubaro PL (2009a) Probing evolutionary patterns in neotropical birds through DNA barcodes. PLoS ONE 4:e4379
Kerr KCR, Birks SM, Kalyakin MV, Red’kin YA, Koblik EA, Hebert PDN (2009b) Filling the gap—COI barcode resolution in eastern Palearctic Birds. Front Zool 6:29
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kruckenhauser L, Haring E, Pinsker W, Riesing MJ, Winkler H, Wink M, Gamauf A (2004) Genetic versus morphological differentiation of Old World buzzards (genus Buteo, Accipitridae). Zool Scripta 33:197–211
Lefébure T, Douady CJ, Gouy M, Gilbert J (2006) Relationship between morphological taxonomy and molecular divergence within crustacea: proposal of a molecular threshold to help species delimitation. Mol Phylogen Evol 40:435–447
Lerner HRL (2007) Molecular phylogenetics of diurnal birds of prey in the avian Accipitridae family (dissertation). University of Michigan, Ann Arbor
Lerner HRL, Mindell DP (2005) Phylogeny of eagles, old world vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Mol Phyl Evol 37:327–346
Lerner HRL, Klaver MC, Mindell DP (2008) Molecular phylogenetics of the Buteonine birds of prey (Accipitridae). Auk 304:304–315
Louette M (2002) Relationships of the red-thighed sparrowhawk Accipiter erythropus and the African Little Sparrowhawk A. minullus. Bull Brit Ornith Club 122:218–222
Louette M (2003) The endemic Ethiopian race of the African Goshawk. Bull Afr Bird Club 10:118–119
Louette M (2007a) Comparative biology of the forest-inhabiting hawks Accipiter spp. in the Democratic Republic of Congo. Ostrich 78:21–28
Louette M (2007b) The variable morphology of the African goshawk (Accipiter tachiro). Ostrich 78:387–393
Louette M, Herremans M (1985) Taxonomy and evolution in the bulbuls (Hypsipetes) on the Comoro Islands. In: Proc Int Symp African Vertebrates, Bonn, West Germany, 15–18 May 1984, pp 407–423
Louette M, Herremans M, Nagy ZT, René de Roland L-A, Jordaens K, Van Houdt J, Sonet G, Breman FC (2011) Frances’ Sparrowhawk Accipiter francesiae (Aves: Accipitridae) radiation on the Comoro Islands. Bonner Zool Monogr 57:133–143
Mayr E (1949) Geographical variation in Accipiter trivirgatus. Am Mus Novitat 1415:1–12
Mayr G, Manegold A, Johansson US (2003) Monophyletic groups within “higher land birds”—comparison of morphological and molecular data. J Zool Syst Evol Res 41:233–248
Meier CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3:e422
Meier R, Shiyang K, Vaidya G, Ng PKL (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55:715–728
Meusnier I, Singer GAC, Landry J-F, Hickey DA, Hebert PDN, Hajibabei M (2008) A universal DNA mini-barcode for biodiversity analysis. BMC Genom 9:214
Ong PS, Luczon AU, Quilang JP, Sumaya AM, Ibañez JC, Salvador DJ, Fontanilla IKC (2011) DNA barcoding of Philippine accipitrids. Mol Ecol Res 11:245–254
Pagès M, Chaval Y, Herbretaus V, Waengsothorn S, Cosson J-F, Hugot J-P, Morand S, Michaux J (2010) Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitatation of species boundaries. BMC Evol Biol 10:184
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
Rach J, DeSalle R, Sarkar IN, Schierwater B, Hadrys H (2008) Character based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc R Soc Lond B 275:237–247
Rambaut A, Drummond AJ (2009) Tracer v1.5. Available from http://beast.bio.ed.ac.uk/Tracer
Ranker TA, Geiger JMO, Kennedy SC, Smith AR, Haufler CH, Paris BS (2003) Molecular phylogenetics and evolution of the endemic Hawaiian genus Adenophorus (Grammitidaceae). Mol Phylogen Evol 26:337–347
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecol Notes 7:355–364
Reynard GB, Short LL, Garrido OH, Alayón G (1987) Nesting, voice, status, and relationships of the endemic Cuban Gundlach’s Hawk (Accipiter gundlachi). Wilson Bull 99:73–77
Riegner MF (2008) Parallel evolution of plumage pattern and coloration in birds: implications for defining avian morphospace. Condor 110:599–614
Rodríguez-Santana F (2010) Reports of Cooper’s hawks (Accipiter cooperii), Swainson’s hawk (Buteo swainsoni) and Short-tailed hawks (Buteo brachyurus) in Cuba. J Raptor Res 44:146–150
Ronquist F, Huelsenbeck JP (2003) MrBayes3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ross HA, Murugan S, Sibon Li WL (2008) Testing the reliability of genetic methods of species identification via simulation. Syst Biol 57:216–230
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Snow DW (1978) An atlas of speciation of African non-passerine birds. Trustees of the British Museum (Natural History), London
Sonet G, Breman FC, Jordaens K, Lenglet G, Louette M, Montañés G, Nagy ZT, Van Houdt J, Verheyen E (2011) Applicability of DNA barcoding to museum specimens of birds from the Democratic Republic of the Congo. Bonner Zool Monogr 57:117–131
Stresemann E (1923) Ueber einige Accipiter-Arten. Journal für Ornithologie 71:517–525
Stresemann E, Amadon D (1979) Falconiformes. In: Mayr E, Cottrell GW (eds) Check-list of birds of the world, vol 1, 2nd edn. Museum of Comparative Zoology, Cambridge, pp 274–425
Swann HK (1922) A synopsis of the Accipitres (diurnal birds of prey), 2nd edn. Wheldon & Wesley, London
Swofford DL (2002) PAUP* phylogenetic analysis using parsimony (*and other methods), version 4b10. Sinauer Associates, Sunderland
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tavares ES, Baker AJ (2008) Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol Biol 8:81
Thiollay SM (1994) Accipiter. In: del Hoyo J, Elliott A, Sargatal JP (eds) Handbook of the birds of the world, New World vultures to guineafowl, vol 2. Lynx, Barcelona
Vaurie C (1961) Systematic notes on palearctic birds. No. 46. Accipitridae: the genus Accipiter. Am Mus Novitat 2039:1–10
Virgilio M, Backeljau T, Nevado B, De Meyer M (2010) Comparative performances of DNA barcoding across insect orders. BMC Bioinf 11:206
Ward RD, Costa FO, Holmes BH, Steinke D (2008) DNA barcoding shared fish species from the North Atlantic and Australasia: minimal divergence for most taxa but likely two species for Zeus faber (john dory) and Lepidopus caudatus (silver scabbardfish). Aquat Biol 3:71–78
Wattel J (1973) Geographical differentiation in the genus Accipiter. Publ Nuttall Ornithol Club 13:1–231
Wink M, Sauer-Gürth H (2000) Advances in molecular systematics of African raptors. In: Chancellor RD, Meyburg BU (eds) Raptors at risk. WWGBP/Hancock House, Berlin, pp 135–147
Wink M, Sauer-Gürth H (2004) Phylogenetic relationships in diurnal raptors based on nucleotide sequences of mitochondrial and nuclear marker genes. In: Chancellor RD, Meyburg BU (eds) Raptors at risk. WWGBP/Hancock House, Berlin, pp 483–498
Yassin A, Amedegnato C, Cruaud C, Veuille M (2009) Molecular taxonomy and species delimitation in Andean Schistocerca (Orthoptera: Acrididae). Mol Phylogen Evol 53:404–411
Yoo HS, Eah J-Y, Kim JS, Kim Y-J, Min M-S, Paek WK, Lee H, Kim C-B (2006) DNA barcoding Korean birds. Mol Cells 22:323–327
Yosef R, Helbig AJ, Clark WS (2001) An intrageneric Accipiter hybrid from Eilat, Israel. Sandgrouse 23:141–144
Acknowledgments
This work would have been impossible without the help of many people we would like to thank: everybody who collected samples, cut toe pads, identified specimens, and kindly allowed us to access their collections. We thank Gerhard Aubrecht (Ober-Österreichische Landesmuseen, Linz Austria), Marieke Berkvens (Wuustwezel, Belgium), Charles Botha (South Africa), Sebastien Bruaux (RBINS), Kizungu Byamana (Lwiro, DR Congo), Chang Yuong Choi (Migratory Bird Centre of the Korean National Park Service, South Korea), Stijn Cooleman (RMCA), S. Viñas & Lellani Fariñes Crespo (RMCA), Réné De Roland Lily Arison (Peregrine Fund, Madagascar), Renate van den Elzen (Zoologisches Forschungsmuseum Koenig, Bonn, Germany), Clem Fisher (World Museum, Liverpool, UK), Jon Fjeldså (University of Copenhagen Zoological Museum, Denmark), Marc Herremans (Mechelen, Belgium), Jon Bolding Kristensen (University of Copenhagen Zoological Museum, Denmark), Georges Lenglet (RBINS), Danny Meirte (RMCA), Jürgen Plass (Ober-Österreichische Landesmuseen, Linz, Austria), Alain Reygel (RMCA), Lucia Liu Severinghaus (The Biodiversity Research Museum Taiwan, Taiwan), Erik Verheyen (RBINS), Malcolm Wilson (South Africa), and Reuven Yosef (International Birding & Research Centre, Eilat, Israel). We further thank two anonymous referees for their valuable comments. All experiments complied with the laws of Belgium.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wink.
Electronic supplementary material
Below is the link to the electronic supplementary material.



Appendices
Appendix 1
See Table 2.
Appendix 2
See Table 3.
Appendix 3
See Fig. 6.
Some examples of nucleotide character that were diagnostic for a species (sensu Rach et al. 2008). The top row of numbers gives the bp postions in the aligned 647 bp barcode fragment. The bottom row indicates whether the given position was located at the first, second, or third codon position
Rights and permissions
About this article
Cite this article
Breman, F.C., Jordaens, K., Sonet, G. et al. DNA barcoding and evolutionary relationships in Accipiter Brisson, 1760 (Aves, Falconiformes: Accipitridae) with a focus on African and Eurasian representatives. J Ornithol 154, 265–287 (2013). https://doi.org/10.1007/s10336-012-0892-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-012-0892-5