Abstract
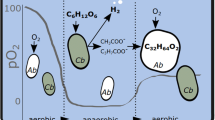
Massive emission of CO2 into atmosphere from consumption of carbon deposit is causing climate change. Researchers have applied metabolic engineering and synthetic biology techniques for improving CO2 fixation efficiency in many species. One solution might be the utilization of autotrophic bacteria, which have great potential to be engineered into microbial cell factories for CO2 fixation and the production of chemicals, independent of fossil resources. In this work, several pathways of Ralstonia eutropha H16 were modulated by manipulation of heterologous and endogenous genes related to fatty acid synthesis. The resulting strain B2(pCT, pFP) was able to produce 124.48 mg/g (cell dry weight) free fatty acids with fructose as carbon source, a fourfold increase over the parent strain H16. To develop a truly autotrophic fermentation technique with H2, CO2 and O2 as substrates, we assembled a relatively safe, continuous, lab-scale gas fermentation system using micro-fermentation tanks, H2 supplied by a hydrogen generator, and keeping the H2 to O2 ratio at 7:1. The system was equipped with a H2 gas alarm, rid of heat sources and placed into a fume hood to further improve the safety. With this system, the best strain B2(pCT, pFP) produced 60.64 mg free fatty acids per g biomass within 48 h, growing in minimal medium supplemented with 9 × 103 mL/L/h hydrogen gas. Thus, an autotrophic fermentation technique to produce fatty acids was successfully established, which might inspire further research on autotrophic gas fermentation with a safe, lab-scale setup, and provides an alternative solution for environmental and energy problems.



Similar content being viewed by others
Change history
10 April 2019
The article Development of an autotrophic fermentation technique for the production of fatty acids using an engineered
References
Cox PM, Betts RA, Jones CD, Spall SA, Totterdell IJ (2000) Acceleration of global warming due to carbon-cycle feedbacks in a coupled climate model. Nature 408:184–187. https://doi.org/10.1038/35041539
Quadrelli Elsje Alessandra, Centi Gabriele, Duplan Jean-Luc, Perathoner Siglinda, Quadrelli EA, Centi G, Duplan JL, Perathoner S (2011) Carbon dioxide recycling: emerging large-scale technologies with industrial potential. Chemsuschem 4:1194–1215. https://doi.org/10.1002/cssc.201100473
Cai Z, Liu G, Zhang J, Li Y (2014) Development of an activity-directed selection system enabled significant improvement of the carboxylation efficiency of Rubisco. Protein Cell 5:552–562. https://doi.org/10.1007/s13238-014-0072-x
Myat T, Lin AO, John Andralojc P, Parry Martin A J, Hanson Maureen R (2014) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513:547–550. https://doi.org/10.1038/nature13776
Chakravarty J, Brigham CJ (2018) Solvent production by engineered Ralstonia eutropha: channeling carbon to biofuel. Appl Microbiol Biotechnol 102:5021–5031. https://doi.org/10.1007/s00253-018-9026-1
Cramm R (2009) Genomic view of energy metabolism in Ralstonia eutropha H16. J Mol Microbiol Biotechnol 16:38–52. https://doi.org/10.1159/000142893
Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voß I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B (2006) Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24:1257–1262. https://doi.org/10.1038/nbt1244
Schwartz E, Henne A, Cramm R, Eitinger T, Friedrich B, Gottschalk G (2003) Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based Lithoautotrophy and Anaerobiosis. J Mol Biol 332:369–383. https://doi.org/10.1016/s0022-2836(03)00894-5
Ishizaki A, Taga N (2001) Microbial production of poly-d-3-hydroxybutyrate from CO2. Appl Microbiol Biotechnol 57:6–12. https://doi.org/10.1007/s002530100775
Reinecke F (2009) Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol 16:91–108. https://doi.org/10.1159/000142897
Shota Atsumi TH, Liao James C (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. https://doi.org/10.1038/nature06450
William B, Black LZ, Kamoku Cody, Liao James C, Li Han (2018) Rearrangement of coenzyme A-acylated carbon chain enables synthesis of isobutanol via a novel pathway in Ralstonia eutropha. ACS Synth Biol 7:794–800. https://doi.org/10.1021/acssynbio.7b00409
Müller Jana, MacEachran Daniel, Burd Helcio, Beller Harry R (2013) Engineering of Ralstonia eutropha H16 for autotrophic and heterotrophic production of methyl ketones. Appl Environ Microbiol 79:4433–4439. https://doi.org/10.1128/AEM.00973-13
Bi Changhao, Peter Su, Müller Jana, Yeh Yi-Chun, Chhabra Swapnil R, Beller Harry R, Singer Steven W, Hillson Nathan J (2013) Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microb Cell Fact 12:1–10. https://doi.org/10.1186/1475-2859-12-107
Tanaka Kenji, Ishizaki Ayaaki, Kanamaru Toshihisa, Kawano Takeharu (1994) Production of poly(d-3-hydroxybutyrate) from CO2, H2, and O2 by high cell density autotrophic cultivation of Alcaligenes eutrophus. Biotechnol Bioengineering 45:268–275. https://doi.org/10.1002/bit.260450312
Handke P, Lynch SA, Gill RT (2011) Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng 13:28. https://doi.org/10.1016/j.ymben.2010.10.007
Gibson Daniel G, Young Lei, Ray-Yuan Chuang J, Venter Craig, Hutchison Clyde A, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. https://doi.org/10.1038/nmeth.1318
Quan J, Tian J (2011) Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc 6:242–251. https://doi.org/10.1038/nprot.2010.181
Hillson Nathan J, Rosengarten Rafael D, Keasling Jay D (2012) j5 DNA assembly design automation software. ACS Synth Biol 1:14–21. https://doi.org/10.1021/sb2000116
Tung T, Hoang RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. https://doi.org/10.1016/s0378-1119(98)00130-9
Amusquivar E, Schiffner S, Herrera E (2011) Evaluation of two methods for plasma fatty acid analysis by GC. Eur J Lipid Sci Technol 113:711–716. https://doi.org/10.1002/ejlt.201000476
Guo D, Zhu J, Deng Z, Liu T (2014) Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways. Metab Eng 22:69–75. https://doi.org/10.1016/j.ymben.2014.01.003
Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA (2013) Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 4:1409. https://doi.org/10.1038/ncomms2425
Zaigao Tan JMY, Chowdhury Anupam, Burdick Kaitlin, Jarboe Laura R, Maranas Costas D, Shanks Jacqueline V (2018) Engineering of E. coli inherent fatty acid biosynthesis capacity to increase octanoic acid production. Biotechnol Biofuels 11:1. https://doi.org/10.1186/s13068-018-1078-z
Tang X, Feng H, Chen WN (2013) Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab Eng 16:95–102. https://doi.org/10.1016/j.ymben.2013.01.003
Li H, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC (2012) Integrated electromicrobial conversion of CO2 to higher alcohols. Science 335:1596. https://doi.org/10.1126/science.1217643
Bredwell Marshall D, Worden RM (1999) Reactor design issues for synthesis-gas fermentations. Biotechnol Prog 15:834–844. https://doi.org/10.1021/bp990108m
Toshikazu Sugimoto TT, Tanaka Kenji, Ishizaki Ayaaki (1999) Control of acetic acid concentration by ph-stat continuous substrate feeding in heterotrophic culture phase of two-stage cultivation of Alcaligenes eutrophus for production of P(3HB) from CO2, H2, and O2 under non-explosive conditions. Biotechnol Bioeng 62:625–631. https://doi.org/10.1002/(SICI)1097-0290(19990320)62:6%3c625:AID-BIT1%3e3.0.CO;2-D
Kodama T, Igarashi Y, Minoda Y (2014) Isolation and culture conditions of a bacterium grown on hydrogen and carbon dioxide. Agric Biol Chem 39:77–82. https://doi.org/10.1080/00021369.1975.10861578
Goh E-B, Baidoo EEK, Keasling Jay D, Beller Harry R (2011) Engineering of bacterial methyl ketone synthesis for biofuels. Appl Environ Microbiol 78:70–80. https://doi.org/10.1128/aem.06785-11
Marc J, Grousseau E, Lombard E, Sinskey AJ, Gorret N, Guillouet SE (2017) Over expression of GroESL in Cupriavidus necator for heterotrophic and autotrophic isopropanol production. Metab Eng 42:74–84. https://doi.org/10.1016/j.ymben.2017.05.007
Schwartz E, Voigt B, Zuhlke D, Pohlmann A, Lenz O, Albrecht D, Schwarze A, Kohlmann Y, Krause C, Hecker M, Friedrich B (2009) A proteomic view of the facultatively chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics 9:5132–5142. https://doi.org/10.1002/pmic.200900333
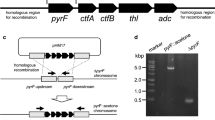
Simon R, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. https://doi.org/10.1038/nbt1183-784
Acknowledgements
This research was financially supported by the Key Research Program of the Chinese Academy of Science (KFZD-SW-215, ZDRW-ZS-2016-3), National Natural Science Foundation of China (31522002, 31770105).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: Due to open choice cancellation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Xiong, B., Liu, L. et al. Development of an autotrophic fermentation technique for the production of fatty acids using an engineered Ralstonia eutropha cell factory. J Ind Microbiol Biotechnol 46, 783–790 (2019). https://doi.org/10.1007/s10295-019-02156-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-019-02156-8