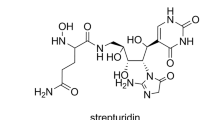
Abstract
Pseudouridimycin (PUM) is a novel pseudouridine-containing peptidyl-nucleoside antibiotic that inhibits bacterial RNA polymerase (RNAP) through a binding site and mechanism different from those of clinically approved RNAP inhibitors of the rifamycin and lipiarmycin (fidaxomicin) classes. PUM was discovered by screening microbial fermentation extracts for RNAP inhibitors. In this review, we describe the discovery and characterization of PUM. We also describe the RNAP-inhibitory and antibacterial properties of PUM. Finally, we review available information on the gene cluster and pathway for PUM biosynthesis and on the potential for discovering additional novel pseudouridine-containing nucleoside antibiotics by searching bacterial genome and metagenome sequences for sequences similar to pumJ, the pseudouridine-synthase gene of the PUM biosynthesis gene cluster.


(Figure adapted from Ref. [26])

(Figure from Ref. [26])

(Figure adapted from Ref. [37])

Similar content being viewed by others
References
Amirkia V, Heinrich M (2015) Natural products and drug discovery: a survey of stakeholders in industry and academia. Front Pharmacol 6:237
Bae B, Nayak D, Ray A, Mustaev A, Landick R, Darst S (2015) CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition. Proc Natl Acad Sci USA 112:E4178–E4187
Belogurov G et al (2009) Transcription inactivation through local refolding of the RNA polymerase structure. Nature 45:332–335
Brown E, Wright G (2016) Antibacterial drug discovery in the resistance era. Nature 529:336–343
Butler MS, Blaskovich MA, Cooper MA (2017) Antibiotics in the clinical pipeline at the end of 2015. J Antibiot 70:3–24
Campbell E, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst S (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912
David B, Wolfender J-L, Dias DA (2015) The pharmaceutical industry and natural products: historical status and new trends. Phytochem Rev 14:299–315
Degen D et al (2014) Transcription inhibition by the depsipeptide antibiotic salinamide A. eLife 3:e02451
Ebright RH (2017) Novel RNA polymerase inhibitor found in soil extracts provides hope for future antibacterial drugs. Future Med Chem 9:1857–1861
Ericsson UB, Andersson ME, Engvall B, Nordlund P, Hallberg BM (2004) Expression, purification, crystallization and preliminary diffraction studies of the tRNA pseudouridine synthase TruD from Escherichia coli. Acta Crystallogr D Biol Crystallogr 60:775–776
Feng Y et al (2015) Structural basis of transcription inhibition by CBR hydroxamidines and CBR pyrazoles. Structure 23:1470–1481
Fernandes P (2015) The global challenge of new classes of antibacterial agents: an industry perspective. Curr Opin Pharmacol 24:7–11
Garibyan L et al (2003) Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2:593–608
Genilloud O (2017) Actinomycetes: still a source of novel antibiotics. Nat Prod Rep 34:1203–1232
Hamma T, Ferré-D’Amaré AR (2006) Pseudouridine synthases. Chem Biol 13:1125–1135
Ho MX, Hudson BP, Das K, Arnold E, Ebright RH (2009) Structures of RNA polymerase–antibiotic complexes. Curr Opin Struct Biol 19:715–723
Holowachuk SA, Bal’a MF, Buddington RK (2003) A kinetic microplate method for quantifying the antibacterial properties of biological fluids. J Microbiol Methods 55:441–446
Iorio M et al (2014) A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol 9:398–404
Jin DJ, Gross C (1988) Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58
Jovanovic M, Burrows P, Bose D, Cámara B, Wiesler S, Weinzierl R, Zhang X, Wigneshweraraj S, Buck M (2011) An activity map of the Escherichia coli RNA polymerase bridge helix. J Biol Chem 286:14469–14479
Li Y, Zhong Z, Zhang W, Qian P-Y (2018) Discovery of cationic nonribosomal peptides as Gram-negative antibiotics through global genome mining. Nat Comm 9:3273
Lin W et al (2017) Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Mol Cell 66:169–179
Lin W et al (2018) Structural basis of transcription inhibition by fidaxomicin (lipiarmycin A3). Mol Cell 70:60–71
Ma C, Yang X, Lewis PJ (2016) Bacterial transcription as a target for antibacterial drug development. Microbiol Mol Biol Rev 80:139–160
Maffioli SI, Cruz JC, Monciardini P, Sosio M, Donadio S (2016) Advancing cell wall inhibitors towards clinical applications. J Ind Microbiol Biotechnol 43:177–184
Maffioli SI et al (2017) Antibacterial nucleoside-analog inhibitor of bacterial RNA polymerase. Cell 169:1240–1248
Maio A, Brandi L, Donadio S, Gualerzi CO (2016) The oligopeptide permease Opp mediates illicit transport of the bacterial P-site decoding inhibitor GE81112. Antibiotics 5:E17
Monciardini P, Iorio M, Maffioli S, Sosio M, Donadio S (2014) Discovering new bioactive compounds from microbial sources. Microb Biotechnol 7:209–220
Mukhopadhyay J et al (2008) The RNA polymerase “switch region” is a target for inhibitors. Cell 135:295–307
National Committee for Clinical Laboratory Standards (2010) Performance standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement—CLSI document M100-S20 (2010) NCCLS. Wayne, Pennsylvania
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661
Niu G, Tan H (2015) Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol 23:110–119
Pesic A, Steinhaus B, Kemper S, Nachtigall J, Kutzner HJ, Höfle G, Süssmuth RD (2014) Isolation and structure elucidation of the nucleoside antibiotic strepturidin from Streptomyces albus DSM 40763. J Antibiot 67:471–477
Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG (2017) Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA 114:5601–5606
Sagitov V, Nikiforov V, Goldfarb A (1993) Dominant lethal mutations near the 5′ substrate binding site affect RNA polymerase propagation. J Biol Chem 268:2195–2202
Silver LL (2016) Natural products as a source of drug leads to overcome drug resistance. Future Microbiol 10:1711–1718
Sosio M, Gaspari E, Iorio M, Pessina S, Medema MH, Bernasconi A, Simone M, Maffioli SI, Ebright RH, Donadio S (2018) Analysis of the pseudouridimycin biosynthetic pathway provides insights into the formation of C-nucleoside antibiotics. Cell Chem Biol 25:540–549
Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A (2005) The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucl Acids Res 33:4202–4211
Srivastava A et al (2011) New target for inhibition of bacterial RNA polymerase: “switch region”. Curr Opin Microbiol 14:532–543
Svetlov V, Vassylyev D, Artsimovitch I (2004) Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J Biol Chem 279:38087–38090
Temiakov D et al (2005) Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell 19:655–666
Tuske S et al (2005) Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 122:541–552
Winn M, Goss RJ, Kimura K, Bugg TD (2010) Antimicrobial antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep 27:279–304
Wright GD (2017) Opportunities for natural products in 21st century antibiotic discovery. Nat Prod Rep 34:694–701
Yuzenkova Y, Roghanian M, Zenkin N (2012) Multiple active centers of multi-subunit RNA polymerases. Transcription 3:115–118
Zhang Y et al (2014) GE23077 binds to the RNA polymerase ‘i’ and ‘i + 1’ sites and prevents the binding of initiating nucleotides. eLife 3:e02450
Acknowledgements
This work was partially supported by NIH Grants GM041376 and AI104660 to RHE and partially supported from a Grant from MIUR Regione Lombardia to NAICONS. We are grateful to all coauthors of the two papers that provided the material for this review.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Special Issue “Natural Product Discovery and Development in the Genomic Era 2019”.
Rights and permissions
About this article
Cite this article
Maffioli, S.I., Sosio, M., Ebright, R.H. et al. Discovery, properties, and biosynthesis of pseudouridimycin, an antibacterial nucleoside-analog inhibitor of bacterial RNA polymerase. J Ind Microbiol Biotechnol 46, 335–343 (2019). https://doi.org/10.1007/s10295-018-2109-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2109-2