Abstract
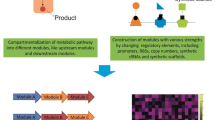
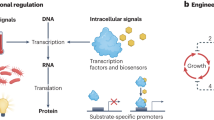
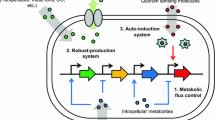
Advances in metabolic engineering have led to the synthesis of a wide variety of valuable chemicals in microorganisms. The key to commercializing these processes is the improvement of titer, productivity, yield, and robustness. Traditional approaches to enhancing production use the “push–pull-block” strategy that modulates enzyme expression under static control. However, strains are often optimized for specific laboratory set-up and are sensitive to environmental fluctuations. Exposure to sub-optimal growth conditions during large-scale fermentation often reduces their production capacity. Moreover, static control of engineered pathways may imbalance cofactors or cause the accumulation of toxic intermediates, which imposes burden on the host and results in decreased production. To overcome these problems, the last decade has witnessed the emergence of a new technology that uses synthetic regulation to control heterologous pathways dynamically, in ways akin to regulatory networks found in nature. Here, we review natural metabolic control strategies and recent developments in how they inspire the engineering of dynamically regulated pathways. We further discuss the challenges of designing and engineering dynamic control and highlight how model-based design can provide a powerful formalism to engineer dynamic control circuits, which together with the tools of synthetic biology, can work to enhance microbial production.


Similar content being viewed by others
References
Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461
Anesiadis N, Cluett WR, Mahadevan R (2008) Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng 10:255–266
Ang J, Harris E, Hussey BJ, Kil R, McMillen DR (2013) Tuning response curves for synthetic biology. ACS Synth Biol 2:547–567
Aström KJ, Murray RM (2008) Feedback systems: an introduction for scientists and engineers. Princeton University Press, New Jersey
Baird NJ, Kulshina N, Ferré-D’Amaré AR (2010) Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol 7:328–332
Beisel CL, Smolke CD (2009) Design principles for riboswitch function. PLoS Comput Biol 5:e1000363
Berens C, Groher F, Suess B (2015) RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression. Biotechnol J 10:246–257
Borkowski O, Ceroni F, Stan G-B, Ellis T (2016) Overloaded and stressed: whole-cell considerations for bacterial synthetic biology. Curr Opin Microbiol 33:123–130
Brewster RC, Jones DL, Phillips R (2012) Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput Biol 8:e1002811
Brophy JAN, Voigt CA (2014) Principles of genetic circuit design. Nat Methods 11:508–520
Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84:647–657
Cardinale S, Arkin AP (2012) Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol J 7:856–866
Chappell J, Westbrook A, Verosloff M, Lucks J (2017) Computational design of Small Transcription Activating RNAs (STARs) for versatile and dynamic gene regulation. Nat Commun 8:1051
Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M (2002) Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol Bioeng 79:53–73
Chin C-S, Chubukov V, Jolly ER, DeRisi J, Li H (2008) Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways. PLoS Biol 6:e146
Chubukov V, Gerosa L, Kochanowski K, Sauer U (2014) Coordination of microbial metabolism. Nat Rev Microbiol 12:327–340
Chubukov V, Zuleta IA, Li H (2012) Regulatory architecture determines optimal regulation of gene expression in metabolic pathways. Proc Natl Acad Sci USA 109:5127–5132
Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD (2013) Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 31:1039–1046
Del Vecchio D, Dy AJ, Qian Y (2016) Control theory meets synthetic biology. J R Soc Interface 13(120). https://doi.org/10.1098/rsif.2016.0380
Delvigne F, Zune Q, Lara AR, Al-Soud W, Sørensen SJ (2014) Metabolic variability in bioprocessing: implications of microbial phenotypic heterogeneity. Trends Biotechnol 32:608–616
Dunlop MJ, Keasling JD, Mukhopadhyay A (2010) A model for improving microbial biofuel production using a synthetic feedback loop. Syst Synth Biol 4:95–104
Farmer WR, Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol 18:533–537
Glick BR (1995) Metabolic load and heterologous gene expression. Biotechnol Adv 13:247–261
Gupta A, Reizman IMB, Reisch CR, Prather KLJ (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35:273–279
Huang B, Lu M, Jia D, Ben-Jacob E, Levine H, Onuchic JN (2017) Interrogating the topological robustness of gene regulatory circuits by randomization. PLoS Comput Biol 13:e1005456
Jang S, Jang S, Xiu Y, Kang TJ, Lee S-H, Koffas MAG, Jung GY (2017) Development of artificial riboswitches for monitoring of naringenin in vivo. ACS Synth Biol 6(11):2077–2085
Kadir T, Mannan AA, Kierzek AM, McFadden J, Shimizu K (2010) Modeling and simulation of the main metabolism in Escherichia coli and its several single-gene knockout mutants with experimental verification. Microb Cell Fact 9:88
Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ (2014) Stochasticity of metabolism and growth at the single-cell level. Nature 514:376–379
Kochanowski K, Sauer U, Chubukov V (2013) Somewhat in control–the role of transcription in regulating microbial metabolic fluxes. Curr Opin Biotechnol 24:987–993
Kochanowski K, Sauer U, Noor E (2015) Posttranslational regulation of microbial metabolism. Curr Opin Microbiol 27:10–17
Kotte O, Volkmer B, Radzikowski JL, Heinemann M (2014) Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol Syst Biol 10:736
Kurata H, Sugimoto Y (2018) Improved kinetic model of Escherichia coli central carbon metabolism in batch and continuous cultures. J Biosci Bioeng 125(2):251–257
Labhsetwar P, Cole JA, Roberts E, Price ND, Luthey-Schulten ZA (2013) Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proc Natl Acad Sci 110:14006–14011
Liao C, Blanchard AE, Lu T (2017) An integrative circuit–host modelling framework for predicting synthetic gene network behaviours. Nat Microbiol 2:1658–1666
Liu D, Evans T, Zhang F (2015) Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng 31:35–43
Liu D, Hoynes-O’Connor A, Zhang F (2013) Bridging the gap between systems biology and synthetic biology. Front Microbiol 4:211
Liu D, Xiao Y, Evans BS, Zhang F (2015) Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol 4:132–140
Liu D, Zhang F (2018) Metabolic feedback circuits provide rapid control of metabolite dynamics. ACS Synth Biol. https://doi.org/10.1021/acssynbio.7b00342
Lo T-M, Chng SH, Teo WS, Cho H-S, Chang MW (2016) A two-layer gene circuit for decoupling cell growth from metabolite production. Cell Syst 3:133–143
Mahr R, Frunzke J (2016) Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl Microbiol Biotechnol 100:79–90
Mannan AA, Liu D, Zhang F, Oyarzún DA (2017) Fundamental design principles for transcription-factor-based metabolite biosensors. ACS Synth Biol 6(10):1851–1859
Mannan AA, Toya Y, Shimizu K, McFadden J, Kierzek AM, Rocco A (2015) Integrating kinetic model of E. coli with genome scale metabolic fluxes overcomes its open system problem and reveals bistability in central metabolism. PLoS One 10:e0139507
Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802
Oyarzún DA, Chaves M (2015) Design of a bistable switch to control cellular uptake. J R Soc Interface 12:20150618
Oyarzún DA, Ingalls BP, Middleton RH, Kalamatianos D (2009) Sequential activation of metabolic pathways: a dynamic optimization approach. Bull Math Biol 71:1851–1872
Oyarzún DA, Lugagne J-B, Stan G-BV (2015) Noise propagation in synthetic gene circuits for metabolic control. ACS Synth Biol 4:116–125
Oyarzún DA, Stan G-BV (2013) Synthetic gene circuits for metabolic control: design trade-offs and constraints. J R Soc Interface 10:20120671
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532
Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD (2012) Microbial engineering for the production of advanced biofuels. Nature 488:320–328
Peskov K, Mogilevskaya E, Demin O (2012) Kinetic modelling of central carbon metabolism in Escherichia coli. FEBS J 279:3374–3385
Pisithkul T, Patel NM, Amador-Noguez D (2015) Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol 24:29–37
Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135:216–226
Rode AB, Endoh T, Sugimoto N (2015) Tuning riboswitch-mediated gene regulation by rational control of aptamer ligand binding properties. Angew Chem Int Ed Engl 54:905–909
Schmid JW, Mauch K, Reuss M, Gilles ED, Kremling A (2004) Metabolic design based on a coupled gene expression-metabolic network model of tryptophan production in Escherichia coli. Metab Eng 6:364–377
Schmitz AC, Hartline CJ, Zhang F (2017) Engineering microbial metabolite dynamics and heterogeneity. Biotechnol J 12(10). https://doi.org/10.1002/biot.201700422
Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31:64–68
Shopera T, He L, Oyetunde T, Tang YJ, Moon TS (2017) Decoupling resource-coupled gene expression in living cells. ACS Synth Biol 6:1596–1604
Simeonidis E, Price ND (2015) Genome-scale modeling for metabolic engineering. J Ind Microbiol Biotechnol 42:327–338
Slusarczyk AL, Lin A, Weiss R (2012) Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13:406–420
Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP (2014) Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci USA 111:7427–7432
Soma Y, Hanai T (2015) Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 30:7–15
Stevens JT, Carothers JM (2015) Designing RNA-based genetic control systems for efficient production from engineered metabolic pathways. ACS Synth Biol 4:107–115
Tan C, Marguet P, You L (2009) Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol 5:842–848
Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, Fields S, Church GM, Raman S (2016) Engineering an allosteric transcription factor to respond to new ligands. Nat Methods 13:177–183
van Heerden JH, Wortel MT, Bruggeman FJ, Heijnen JJ, Bollen YJM, Planqué R, Hulshof J, O’Toole TG, Wahl SA, Teusink B (2014) Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343:1245114
Waldherr S, Oyarzún DA, Bockmayr A (2015) Dynamic optimization of metabolic networks coupled with gene expression. J Theor Biol 365:469–485
Wang Y-H, McKeague M, Hsu TM, Smolke CD (2016) Design and construction of generalizable RNA-protein hybrid controllers by level-matched genetic signal amplification. Cell Syst 3(549–562):e547
Weiße AY, Oyarzún DA, Danos V, Swain PS (2015) Mechanistic links between cellular trade-offs, gene expression, and growth. Proc Natl Acad Sci USA 112:E1038–E1047
Xiao Y, Bowen CH, Liu D, Zhang F (2016) Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 12:339–344
Xie W, Ye L, Lv X, Xu H, Yu H (2015) Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng 28:8–18
Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA 111:11299–11304
Xue Z, Sharpe PL, Hong S-P, Yadav NS, Xie D, Short DR, Damude HG, Rupert RA, Seip JE, Wang J, Pollak DW, Bostick MW, Bosak MD, Macool DJ, Hollerbach DH, Zhang H, Arcilla DM, Bledsoe SA, Croker K, McCord EF, Tyreus BD, Jackson EN, Zhu Q (2013) Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol 31:734–740
Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U (2004) Just-in-time transcription program in metabolic pathways. Nat Genet 36:486–491
Zhang F, Carothers JM, Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30:354–359
Zhang F, Keasling J (2011) Biosensors and their applications in microbial metabolic engineering. Trends Microbiol 19:323–329
Zhang J, Jensen MK, Keasling JD (2015) Development of biosensors and their application in metabolic engineering. Curr Opin Chem Biol 28:1–8
Zhou L-B, Zeng A-P (2015) Exploring lysine riboswitch for metabolic flux control and improvement of l-lysine synthesis in Corynebacterium glutamicum. ACS Synth Biol 4:729–734
Acknowledgements
This work was funded by the Human Frontier Science Program through a Young Investigator Grant awarded to D. O. and F. Z. (Grant no. RGY0076-2015) and the US National Science Foundation (MCB1453147) to F. Z.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, D., Mannan, A.A., Han, Y. et al. Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol 45, 535–543 (2018). https://doi.org/10.1007/s10295-018-2013-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2013-9