Abstract
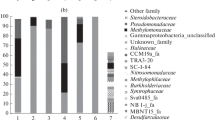
Which bacterial taxonomic groups can be used in monitoring saline water methanol-utilizing denitrification and whether nitrate is transformed into N2 in the process are unclear. Therefore, methylotrophic bacterial communities of two efficiently functioning (nitrate/nitrite reduction was 63–96 %) tropical and cool seawater reactors at a public aquarium were investigated with clone library analysis and 454 pyrosequencing of the 16S rRNA genes. Transformation of nitrate into N2 was confirmed using 15N labeling in incubation of carrier material from the tropical reactor. Combining the data with previous study results, Methylophaga and Hyphomicrobium were determined to be suitable target genera for monitoring the function of saline water methanol-fed denitrification systems. However, monitoring was not possible at the single species level. Interestingly, potential nitrate-reducing methylotrophs within Filomicrobium and closely related Fil I and Fil II clusters were detected in the reactors suggesting that they also contributed to methylotrophic denitrification in the saline environment.




Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Auclair J, Lépine F, Parent S, Villemur R (2010) Dissimilatory reduction of nitrate in seawater by a Methylophaga strain containing two highly divergent narG sequences. ISME J 4:1302–1313
Auclair J, Parent S, Villemur R (2012) Functional diversity in the denitrifying biofilm of the methanol-fed marine denitrification system at the Montreal Biodome. Microb Ecol 63:726–735
Baytshtok V, Lu H, Park H, Kim S, Yu R, Khandran K (2009) Impact of varying electron donors on the molecular microbial ecology and biokinetics of methylotrophic denitrifying bacteria. Biotechnol Bioeng 102:1527–1536
Beck DAC, McTaggart TL, Setboonsarng U, Vorobev A, Kalyuzhnaya MG, Ivanova N, Goodwin L, Woyke T, Lidstrom ME, Chistoserdova L (2014) The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS One 9:e102458
Bernet N, Delgenes N, Akunna JC, Delgenes JP, Moletta R (2000) Combined anaerobic–aerobic SBR for the treatment of piggery wastewater. Water Res 34:611–619
Boden R, Ferriera S, Johnson J, Kelly DP, Murrell JC, Schäfer H (2011) Draft genome sequence of the chemolithoheterotrophic, halophilic methylotroph Methylophaga thiooxydans DMS010. J Bacteriol 193:3154–3155
Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME (2009) The expanding world of methylotrophic organisms. Annu Rev Microbiol 63:477–499
Dernjatin M (2008) Leijuva-alustaisen denitrifikaatioprosessin optimointi suljetussa kiertovesijärjestelmässä. MsC thesis, University of Helsinki, Helsinki, Finland (in Finnish)
Gentile ME, Nyman JL, Criddle CS (2007) Correlation of patterns of denitrification instability in replicated bioreactor communities with shifts in the relative abundance and the denitrification patterns of specific populations. ISME J 1:714–728
Gittel A, Kofoed MVW, Sørensen KB, Ingvorsen K, Schramm A (2012) Succession of Deferribacteres and Epsilonproteobacteria through a nitrate-treated high-temperature oil production facility. Syst Appl Microbiol 35:165–174
Hagman M, Nielsen JL, Nielsen PH, la Jansen J C (2008) Mixed carbon sources for nitrate reduction in activated sludge-identification of bacteria and process activity studies. Water Res 42:1539–1546
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4(1):9
Henriques AC, De Marco P (2015) Complete genome sequences of two strains of “Candidatus Filomicrobium marinum”, a methanesulfonate-degrading species. Genome Announc 3:e00160-15
Hung WL, Wade WG, Boden R, Kelly DP, Wood AP (2011) Facultative methylotrophs from the human oral cavity and methylotrophy in strains of Gordonia, Leifsonia, and Microbacterium. Arch Microbiol 193:407–417
Jensen S, Neufeld JD, Birkeland NK, Hovland M, Murrell JC (2008) Methane assimilation and trophic interactions with marine Methylomicrobium in deep-water coral reef sediment off the coast of Norway. FEMS Microbiol Ecol 66:320–330
Kampschreur MJ, Temmink H, Kleerebezem R, Jetten MSM, van Loosdrecht MCM (2009) Nitrous oxide emission during wastewater treatment. Water Res 43:4093–4103
Koch G, Siegrist H (1997) Denitrification with methanol in tertiary filtration. Water Res 31:3029–3038
Kolb S (2009) Aerobic methanol- oxidizing Bacteria in soil. FEMS Microbiol Lett 300:1–10
Kumaresan D, Wischer D, Hillebrand-Voiculescu AM, Murrell JC (2015) Draft genome sequences of facultative methylotrophs, Gemmobacter sp. strain LW1 and Mesorhizobium sp. strain 1M-11, isolated from Movile Cave, Romania. Genome Announc 3:e01266-15
Labbé N, Juteau P, Parent S, Villemur R (2003) Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal Biodome, Canada. Microb Ecol 46:12–21
Labbé N, Laurin V, Juteau P, Parent S, Villemur R (2007) Microbiological community structure of the biofilm of a methanol-fed, marine denitrification system, and identification of the methanol-utilizing micro-organisms. Microb Ecol 53:621–630
Labbé N, Parent S, Villemur R (2003) Addition of trace metals increases denitrification rate in closed marine systems. Water Res 37:914–920
Labelle M-A, Juteau P, Jolicoeur M, Villemur R, Parent S, Comeau Y (2005) Seawater denitrification in a closed mesocosm by a submerged moving bed biofilm reactor. Water Res 39:3409–3417
Lee PG, Lea RN, Dohmann E, Prebilsky W, Turk PE, Ying H, Whitson JL (2000) Denitrification in aquaculture systems: an example of a fuzzy logic control problem. Aquac Eng 23:37–59
Lemmer H, Zaglauer A, Metzner G (1997) Denitrification in a methanol-fed fixed-bed reactor. Part 1: physico-chemical and biological characterization. Water Res 31:1897–1902
Lu H, Chandran K, Stensel D (2014) Microbial ecology of denitrification in biological wastewater treatment. Water Res 64:237–254
Madhaiyan M, Poonguzhali S, Lee J-S, Lee KC, Sundaram S (2010) Flavobacterium glycines sp. nov., a facultative methylotroph isolated from the rhizosphere of soybean. Int J Syst Evol Microbiol 60:2187–2192
Martineau C, Villeneuve C, Mauffrey F, Villemur R (2013) Hyphomicrobium nitrativorans sp. nov., isolated from the biofilm of a methanol-fed denitrification system treating seawater at the Montreal Biodome. Int J Syst Evol Microbiol 63:3777–3781
Martineau C, Mauffrey F, Villemur R (2015) Comparative analysis of denitrifying activities of Hyphomicrobium nitrativorans, Hyphomicrobium denitrificans and Hyphomicrobium zavarzinii. Appl Environ Microbiol 81:5003–5014
Neufeld JD, Schäfer H, Cox MJ, Boden R, McDonald IR, Murrell JC (2007) Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1:480–491
Osaka T, Shirotani K, Yoshie S, Tsuneda S (2008) Effects of carbon source on denitrification efficiency and microbial community structure in a saline wastewater treatment process. Water Res 42:3709–3718
Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, Jones RI (2012) Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6:1640–1652
Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC (2002) Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331–2342
Rainey FA, Ward-Rainey N, Gliesche CG, Stackebrandt E (1998) Phylogenetic analysis and intrageneric structure of the genus and the related genus Filomicrobium. Int J Syst Bacteriol 48:635–639
Rissanen AJ, Kurhela E, Aho T, Oittinen T, Tiirola M (2010) Storage of environmental samples for guaranteeing nucleic acid yields for molecular microbiological studies. Appl Microbiol Biotechnol 88:977–984
Scheuner C, Tindall BJ, Lu M, Nolan M, Lapidus A, Cheng JF, Goodwin L, Pitluck S, Huntemann M, Liolios K, Pagani I, Mavromatis K, Ivanova N, Pati A, Chen A, Palaniappan K, Jeffries CD, Hauser L, Land M, Mwrichia R, Rohde M, Abt B, Detter JC, Woyke T, Eisen JA, Markowitz V, Hugenholtz P, Göker M, Kyrpides NC, Klenk HP (2014) Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Stand Genomic Sci 9:10
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shoun H, Kano M, Baba I, Takaya N, Matsuo M (1998) Denitrification by Actinomycetes and purification of dissimilatory nitrite reductase and azurin from Streptomyces thioluteus. J Bacteriol 180:4413–4415
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tiirola MA, Rissanen AJ, Sarpakunnas M, Arvola L, Nykänen H (2011) Stable isotope profiles of nitrogen gas indicate denitrification in oxygen-stratified humic lakes. Rapid Commun Mass Spectrom 25:1497–1502
Villeneuve C, Martineau C, Mauffrey F, Villemur R (2013) Methylophaga nitratireducenticrescens sp. nov. and Methylophaga frappieri sp. nov., isolated from the biofilm of the methanol-fed denitrification system treating the seawater at the Montreal Biodome. Int J Syst Evol Microbiol 63:2216–2222
Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H (2002) Microbial community composition and function in wastewater treatment plants. Antonie van Leeuwenhoek 81:665–680
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Countinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, DeBoy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, Gwinn-Giglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren Q, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu C, Zafar N, Zhou L, Kuske CR (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056
Wu X-L, Yu S-L, Gu J, Zhao G-F, Chi C-Q (2009) Filomicrobium insigne sp. nov., isolated from an oil-polluted saline soil. Int J Syst Evol Microbiol 59:300–305
Zhu J, Wang Q, Yuan M, Tan GYA, Sun F, Wang C, Wu W, Lee PH (2016) Microbiology and potential applications of aerobic methane oxidation coupled to denitrification (AME-D) process: a review. Water Res 90:203–215
Acknowledgments
We thank H. Devlin, B. Thamdrup and S. Hallin for comments on an earlier version of this manuscript. This study was funded by Maa-ja Vesitekniikan Tuki ry (A.J.R.) and the Academy of Finland (projects 286642 to A.J.R. and 120089 to M.T.), as well as the European Research Council (ERC) Consolidator project 615146 (M.T.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2016_1839_MOESM1_ESM.pdf
Online Resource 1 Hierarchical clustering analysis (UPGMA) of relative abundance of OTUs (using Bray-Curtis distances) in 16S rRNA gene clone library analyses of AQUAR_T and AQUAR_C in 2008 and 2010 (PDF 13 kb)
Rights and permissions
About this article
Cite this article
Rissanen, A.J., Ojala, A., Dernjatin, M. et al. Methylophaga and Hyphomicrobium can be used as target genera in monitoring saline water methanol-utilizing denitrification. J Ind Microbiol Biotechnol 43, 1647–1657 (2016). https://doi.org/10.1007/s10295-016-1839-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1839-2