Abstract
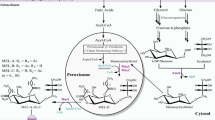
Lipases or triacylglycerol hydrolases are widely spread in nature and are particularly common in the microbial world. The filamentous fungus Mucor circinelloides is a potential lipase producer, as it grows well in triacylglycerol-contained culture media. So far only one lipase from M. circinelloides has been characterized, while the majority of lipases remain unknown in this fungus. In the present study, 47 potential lipase genes in M. circinelloides WJ11 and 30 potential lipase genes in M. circinelloides CBS 277.49 were identified by extensive bioinformatics analysis. An overview of these lipases is presented, including several characteristics, sub-cellular location, phylogenetic analysis and expression profiling of the lipase genes during growth and lipid accumulation. All of these proteins contained the consensus sequence for a classical lipase (GXSXG motif) and were divided into four types including α/β-hydrolase_1, α/β-hydrolase_3, class_3 and GDSL lipase (GDSL) based on gene annotations. Phylogenetic analyses revealed that class_3 family and α/β-hydrolase_3 family were the conserved lipase family in M. circinelloides. Additionally, some lipases also contained a typical acyltransferase motif of H-(X) 4-D, and these lipases may play a dual role in lipid metabolism, catalyzing both lipid hydrolysis and transacylation reactions. The differential expression of all lipase genes were confirmed by quantitative real-time PCR, and the expression profiling were analyzed to predict the possible biological roles of these lipase genes in lipid metabolism in M. circinelloides. We preliminarily hypothesized that lipases may be involved in triacylglycerol degradation, phospholipid synthesis and beta-oxidation. Moreover, the results of sub-cellular localization, the presence of signal peptide and transcriptional analyses of lipase genes indicated that four lipase in WJ11 most likely belong to extracellular lipases with a signal peptide. These findings provide a platform for the selection of candidate lipase genes for further detailed functional study.



Similar content being viewed by others
References
Andrade GS, Carvalho AK, Romero CM, Oliveira PC, Castro HF (2014) Mucor circinelloides whole-cells as a biocatalyst for the production of ethyl esters based on babassu oil. Bioprocess Biosyst Eng 37:2539–2548
Andrade GSS, Freitas L, Oliveira PC, Castro HF (2012) Screening, immobilization and utilization of whole cell biocatalysts to mediate the ethanolysis of babassu oil. J Mol Catal B Enzym 84:183–188
Antczak T (2001) Catalytic properties of fungal lipases from Mucor Circinelloides and Mucor Racemosus. Zeszyty Naukowe. Rozprawy Naukowe/Politechnika Łódzka 298:5–169
Bigey F, Bougard TD, Nicaud JM, Moulin G (2003) Identification of a triacylglycerol lipase gene family in Candida deformans: molecular cloning and functional expression. Yeast 20:233–248
Carvalho AKF, Faria ELP, Rivaldi JD, Andrade GSS, Oliveira PC (2015) Performance of whole-cells lipase derived from Mucor circinelloides as a catalyst in the ethanolysis of non-edible vegetable oils under batch and continuous run conditions. Ind Crops Prod 67:287–294
Chen H, Hao G, Wang L, Wang H, Gu Z (2014) Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci Rep 5:11247. doi:10.1038/srep11247
Desfougeres T, Haddouche R, Fudalej F, Neuveglise C, Nicaud JM (2010) SOA genes encode proteins controlling lipase expression in response to triacylglycerol utilization in the yeast Yarrowia lipolytica. FEMS Yeast Res 10:93–103
Destain J, Fickers P, Weekers F, Moreau B, Thonart P (2005) Utilization of methyloleate in production of microbial lipase. Appl Biochem Biotechnol 121–124:269–277
Dulermo T, Tréton B, Beopoulos A, Gnankon APK, Haddouche R (2013) Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: new insights into the role of intracellular lipases and lipid body organisation. Biochim Biophys Acta 1831:1486–1495
Fickers P, Marty A, Nicaud JM (2011) The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol Adv 29:632–644
Fickers P, Nicaud JM, Gaillardin C, Destain J, Thonart P (2004) Carbon and nitrogen sources modulate lipase production in the yeast Yarrowia lipolytica. J Appl Microbiol 96:742–749
Garcia-Galan C, Barbosa O, Ortiz C, Torres R, Rodrigues RC (2013) Biotechnological prospects of the lipase from Mucor javanicus. J Mol Catal B Enzym 93:34–43
Goncalves C, Lopes M, Ferreira JP, Belo I (2009) Biological treatment of olive mill wastewater by non-conventional yeasts. Bioresour Technol 100:3759–3763
Guieysse D, Sandoval G, Faure L, Nicaud JM, Monsan P (2005) New efficient lipase from Yarrowia lipolytica for the resolution of 2-bromo-arylacetic acid esters. Tetrahedron Asymmetry 15(22):3539–3543
Ham HJ, Rho HJ, Shin SK, Yoon HJ (2010) The TGL2 gene of Saccharomyces cerevisiae encodes an active acylglycerol lipase located in the mitochondria. J Biol Chem 285:3005–3013
Holmes RS, Cox LA, VandeBerg JL (2010) Comparative studies of mammalian acid lipases: evidence for a new gene family in mouse and rat (Lipo). Comp Biochem Physiol Part D Genom Proteom 5:217–226
Kamzolova SV, Finogenova TV, Lunina YN, Perevoznikova OA, Minachova LN (2007) Characteristics of the growth on rapeseed oil and synthesis of citric and isocitric acids by Yarrowia lipolytica yeasts. Microbiology 76:20–24
Kendrick A, Ratledge C (1992) Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur J Biochem 209:667–673
Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K (2006) Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem 281:491–500
Lazniewski M, Steczkiewicz K, Knizewski L, Wawer I, Ginalski K (2011) Novel transmembrane lipases of alpha/beta hydrolase fold. FEBS Lett 585:870–874
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Pleiss J, Scheib H, Schmid RD (2000) The His gap motif in microbial lipases: a determinant of stereoselectivity toward triacylglycerols and analogs. Biochimie 82:1043–1052
Purwanto MGM, Maretha MV, Wahyudi M, Goeltom MT (2015) Whole cell hydrolysis of sardine (Sardinella Lemuru) oil waste using Mucor Circinelloides NRRL 1405 immobilized in poly-urethane foam. Procedia Chem 14:256–262
Rajakumari S, Daum G (2009) Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell 21:501–510
Rajendran A, Palanisamy A, Thangavelu V (2009) Lipase catalyzed ester synthesis for food processing industries. Braz Arch Biol Technol 52:207–219
Rosa Amarilis RF, Adrián G, Santiago TM, Victoriano G (2012) Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl Microbiol Biotechnol 97:3063–3072
Schmidt C, Athenstaedt K, Koch B, Ploier B, Daum G (2013) Regulation of the yeast triacylglycerol lipase TGl3p by formation of nonpolar lipids. J Biol Chem 288:19939–19948
Sona R, Günther D (2010) Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J Biol Chem 285:15769–15776
Suzzi G, Lanorte MT, Galgano F, Andrighetto C, Lombardi A (2001) Proteolytic, lipolytic and molecular characterisation of Yarrowia lipolytica isolated from cheese. Int J Food Microbiol 69:69–77
Szczesna-Antczak M, Antczak T, Piotrowicz-Wasiak M, Rzyska M, Binkowska N, Bielecki S (2006) Relationships between lipases and lipids in mycelia of two Mucor strains. Enzyme Microbial Technol 39:1214–1222
Szczesna-Antczak M, Antczak T, Rzyska M, Modrzejewska Z, Patura J, Kalinowska H, Bielecki S (2004) Stabilization of an intracellular Mucor circinelloides lipase for application in non-aqueous media. J Mol Catal B Enzym 29:163–171
Tang X, Chen HQ, Chen YQ, Chen W, Victoriano G (2015) Comparison of biochemical activities between high and low lipid-producing strains of Mucor circinelloides: an explanation for the high oleaginicity of strain WJ11. PLoS One 10(9):e0137543
Tang X, Zhao LN, Chen HQ, Chen YQ, Chen W, Song YD, Ratledge C (2015) Complete genome sequence of a high lipid-producing strain of Mucor circinelloides WJ11 and comparative genome analysis with a low lipid-producing strain CBS 277.49. PLoS One 10(9):e0137543. doi:10.1371/journal.pone.0137543
Tang X, Zan X, Zhao L, Chen H, Chen YQ, Chen W, Song Y, Ratledge C (2016) Proteomics analysis of high lipid-producing strain Mucor circinelloides WJ11: an explanation for the mechanism of lipid accumulation at the proteomic level. Microb Cell Fact 11:15–35
Turki S, Mrabet JG, Marouani A, Thonart P (2010) Preliminary safety assessment of Yarrowia lipolytica extracellular lipase: results of acute and 28-day repeated dose oral toxicity studies in rats. Food Chem Toxicol 48:2393–2400
Vongsangnak W, Klanchui A, Tawornsamretkit L, Tatiyaborwornchai W, Laoteng K, Meechai A (2016) Genome-scale metabolic modeling of Mucor circinelloides and comparative analysis with other oleaginous species. Gene 583(2):121–129
Yazawa H, Kumagai H, Uemura H (2012) Characterization of triglyceride lipase genes of fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol 96:981–991
Ying Z, Adams IP, Colin R (2007) Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2025
Yoon K, Han D, Li Y, Sommerfeld M, Hu Q (2012) Phospholipid: diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24:3708–3724
Zan XY, Tang X, Zhao LN, Chu LF, Chen HQ, Chen W, Chen YQ, Song YD (2016) Bioinformatical analysis and preliminary study of the role of lipase in lipid metabolism in Mucor circinelloides. RSC Adv 6:60673–60682
Zhao LN, Tang X, Luan X, Chen HQ, Chen YQ, Chen W, Song YD, Ratledge C (2015) Role of pentose phosphate pathway in lipid accumulation of oleaginous fungus Mucor circinelloides. RSC Adv 5:97658–97664
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31271812 and 21276108), the National High Technology Research and Development Program of China (2012AA022105C), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1249), the Program for New Century Excellent Talents (NCET-13-0831).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zan, X., Tang, X., Chu, L. et al. Lipase genes in Mucor circinelloides: identification, sub-cellular location, phylogenetic analysis and expression profiling during growth and lipid accumulation. J Ind Microbiol Biotechnol 43, 1467–1480 (2016). https://doi.org/10.1007/s10295-016-1820-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1820-0