Abstract
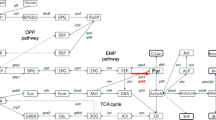
A metabolic network consisting of 48 reactions was established to describe intracellular processes during growth and poly-3-hydroxybutyrate (PHB) production for Cupriavidus necator DSM 545. Glycerol acted as the sole carbon source during exponential, steady-state cultivation conditions. Elementary flux modes were obtained by the program Metatool and analyzed by using yield space analysis. Four sets of elementary modes were obtained, depending on whether the pair NAD/NADH or FAD/FADH2 contributes to the reaction of glycerol-3-phosphate dehydrogenase (GLY-3-P DH), and whether 6-phosphogluconate dehydrogenase (6-PG DH) is present or not. Established metabolic network and the related system of equations provide multiple solutions for the simultaneous synthesis of PHB and biomass; this number of solutions can be further increased if NAD/NADH or FAD/FADH2 were assumed to contribute in the reaction of GLY-3-P DH. As a major outcome, it was demonstrated that experimentally determined yields for biomass and PHB with respect to glycerol fit well to the values obtained in silico when the Entner–Doudoroff pathway (ED) dominates over the glycolytic pathway; this is also the case if the Embden–Meyerhof–Parnas pathway dominates over the ED.






Similar content being viewed by others
References
Braunegg G, Sonnleitner B, Lafferty RM (1978) A rapid gas chromatographic method for the determination of poly-(b-hydroxy-butyric) acid in microbial biomass. Eur J Appl Microbiol Biotechnol 6:29–37
Burgard AP, Nikolaev EV, Schilling CH, Maranas CD (2004) Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res 14:301–312
Bushell ME, Sequeira SIP, Khannapho C, Zhao H, Chater KF, Butler MJ, Kierzek AM, Avignone-Rossa CA (2006) The use of genome scale metabolic flux variability analysis for process feed formulation based on an investigation of the effects of the zwf mutation on antibiotic production in Streptomyces coelicolor. Enzyme Microb Technol 39:1347–1353
Cavalheiro JMBT, Raposo RS, de Almeida MCMD, Cesário MT, Sevrin C, Grandfils C, da Fonseca MMR (2012) Effect of cultivation parameters on the production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) and poly(3-hydroxybutyrate-4-hydroxybutyrate-3-hydroxyvalerate) by Cupriavidus necator using waste glycerol. Bioresour Technol 111:391–397
Chen GQ (2009) A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38:2434–2446
Dräger A, Kronfeld M, Ziller MJ, Supper J, Planatscher H, Magnus JB, Oldiges M, Kohlbacher O, Zell A (2009) Modeling metabolic networks in C. glutamicum: a comparison of rate laws in combination with various parameter optimization strategies. BMC Syst Biol 3:5. doi:10.1186/1752-0509-3-5
Edwards JS, Ramakrishna R, Schilling CH, Palsson BO (1999) Metabolic flux balance analysis. In: Lee SY, Papoutsakis ET (eds) Metabolic engineering. Marcel Dekker, New York, pp 13–57
Fleige C, Kroll J, Steinbüchel A (2011) Establishment of an alternative phosphoketolase-dependent pathway for fructose catabolism in Ralstonia eutropha H16. Appl Microbiol Biotechnol 91:769–776
Franz A, Song HS, Ramkrishna D, Kienle A (2011) Experimental and theoretical analysis of poly(β-hydroxybutyrate) formation and consumption in Ralstonia eutropha. Biochem Eng J 55:49–58
García IL, López JA, Dorado MP, Kopsahelis N, Alexandri M, Papanikolaou S, Villar MA, Koutinas AA (2013) Evaluation of by-products from the biodiesel industry as fermentation feedstock for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) production by Cupriavidus necator. Bioresour Technol 130:16–22
Gombert AK, Nielsen J (2000) Mathematical modelling of metabolism. Curr Opi Biotechnol 11:180–186
Grousseau E, Blanchet E, Déléris S, Albuquerque MG, Paul E, Uribelarrea JL (2013) Impact of sustaining a controlled residual growth on polyhydroxybutyrate yield and production kinetics in Cupriavidus necator. Bioresour Technol 148:30–38
Gudmundsson S, Thiele I (2010) Computationally efficient flux variability analysis. BMC Bioinform 11:489. doi:10.1186/1471-2105-11-489
Kaddor C, Steinbüchel A (2011) Implications of various phosphoenolpyruvate-carbohydrate phosphotransferase system mutations on glycerol utilization and poly(3-hydroxybutyrate) accumulation in Ralstonia eutropha H16, AMB Express. 1:16. Available via http://www.amb-express.com/content/1/1/16
Khanna S, Goyal A, Moholkar VS (2012) Microbial conversion of glycerol: present status and future prospects. Crit Rev Biotechnol 32:235–262
Koller M, Bona R, Braunegg G, Hermann C, Horvat P, Kroutil M, Martinz J, Neto J, Pereira L, Varila P (2005) Production of polyhydroxyalkanoates from agricultural waste and surplus materials. Biomacromolecules 6:561–565
Koller M, Salerno A, Braunegg G (2013) Polyhydroxyalkanoates: basics, production and applications of microbial biopolyesters. In: Kabasci S, Stevens C (eds) Bio-based plastics: materials and applications. Wiley, New York, pp 137–170
König C, Sammler I, Wilde E, Schlegel HG (1969) Konstitutive Glucose-6-phosphat-Dehydrogenase bei Glucose verwertenden Mutanten von einem kryptischen Wildstamm. Arch Mikrobiol 67:51–57
Larhlimi A, Bockmayr A (2009) A new constraint-based description of the steady-state flux cone of metabolic networks. Discrete Appl Math 157:2257–2266
Lee JN, Shin HD, Lee YH (2003) Metabolic engineering of pentose phosphate pathway in Ralstonia eutropha for enhanced biosynthesis of poly-β-hydroxybutyrate. Biotechnol Prog 19:1444–1449
Lopar M, Vrana Špoljarić I, Atlić A, Koller M, Braunegg G, Horvat P (2013) Five-step continuous production of PHB analyzed by elementary flux modes, yield space analysis and high structured metabolic model. Biochem Eng J 79:57–70
Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5:264–276 PMID 14642354
Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nat Biotechnol 28:245–248. doi:10.1038/nbt.1614
Papin JA, Price ND, Palsson BØ (2002) Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome Res 12:1889–1900
Papin JA, Stelling J, Price ND, Klamt S, Schuster S, Palsson BO (2004) Comparison of network-based pathway analysis methods. Trends Biotechnol 22:400–405
Park JM, Kim TY, Lee SY (2010) Prediction of metabolic fluxes by incorporating genomic context and flux-converging pattern analyses. Proc Natl Acad Sci USA 107:14931–14936
Park JM, Kim TY, Lee SY (2011) Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production. BMC Syst Biol 5:101. doi:10.1186/1752-0509-5-101
Pfeiffer T, Sánchez-Valdenebro I, Nuño JC, Montero F, Schuster S (1999) METATOOL: for studying metabolic networks. Bioinformatics 15:251–257
Pöhlein A, Kusian B, Friedrich B, Daniel R, Bowien B (2011) Complete genome sequence of the type strain Cupriavidus necator N-1. J Bacteriol 193:5017
Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B (2006) Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24:1257–1262
Price ND, Reed JL, Papin JA, Wiback SJ, Palsson BO (2003) Network-based analysis of metabolic regulation in the human red blood cell. J Theor Biol 225:185–194
Raberg M, Kaddor C, Kusian B, Stahlhut G, Budinova R, Kolev N, Bowien B, Steinbüchel A (2012) Impact of each individual component of the mutated PTS(Nag) on glucose uptake and phosphorylation in Ralstonia eutropha G+1. Appl Microbiol Biotechnol 95:735–744
Raberg M, Peplinski K, Heiss S, Ehrenreich A, Voigt B, Döring C, Bömeke M, Hecker M, Steinbüchel A (2011) Proteomic and transcriptomic elucidation of the mutant Ralstonia eutropha G+1 with regard to glucose utilization. Appl Environ Microbiol 77:2058–2070
Reinecke F, Steinbüchel A (2009) Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol 16:91–108
Schlegel HG, Gottschalk G (1965) Verwertung von Glucose durch eine Mutante von Hydrogenomonas H16. Biochem Z 341:249–259
Schuster S, Fell DA, Dandekar T (2000) A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat Biotechnol 18:326–332
Schuster S, Hilgetag C (1994) On elementary flux modes in biochemical reaction systems at steady state. J Biol Syst 2:165–182. doi:10.1142/S0218339094000131
Schwartz JM, Kanehisa M (2005) A quadratic programming approach for decomposing steady-state metabolic flux distributions onto elementary modes. Bioinformatics 21:204–205
Schweizer HP, Jump R, Po C (1997) Structure and gene-polypeptide relationships of the region encoding glycerol diffusion facilitator (glpF) and glycerol kinase (glpK) of Pseudomonas aeruginosa. Microbiology 143:1287–1297
Sichwart S, Hetzler S, Bröker D, Steinbüchel A (2011) Extension of the substrate utilization range of Ralstonia eutropha strain H16 by metabolic engineering to include mannose and glucose. Appl Environ Microbiol 77:1325–1334
Song HS, Morgan JA, Ramkrishna D (2009) Systematic development of hybrid cybernetic models: application to recombinant yeast co-consuming glucose and xylose. Biotechnol Bioeng 103:984–1002
Song HS, Ramkrishna D (2009) Reduction of a set of elementary modes using yield analysis. Biotechnol Bioeng 102:554–568
Steinbüchel A (1986) Expression of the Escherichia coli pfkA gene in Alcaligenes eutrophus and in other gram-negative bacteria. J Bacteriol 166:319–327
Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED (2002) Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190–193
Stephanopoulos GN, Aristidou A, Nielsen J (1998) Metabolic engineering: principles and methodologies. Academic Press, San Diego
Varma A, Palsson BØ (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Nat Biotechnol 12:994–998
von Kamp A, Schuster S (2006) Metatool 5.0: fast and flexible elementary modes analysis. Bioinformatics 22:1930–1931
Vrana Špoljarić I, Lopar M, Koller M, Muhr A, Salerno A, Reiterer A, Horvat P (2013) In silico optimization and low structured kinetic model of poly[(R)-3-hydroxybutyrate] synthesis by Cupriavidus necator DSM 545 by fed-batch cultivation on glycerol. J Biotechnol 168:625–635
Vrana Špoljarić I, Lopar M, Koller M, Muhr A, Salerno A, Reiterer A, Malli K, Angerer H, Strohmeier K, Schober S, Mittelbach M, Horvat P (2013) Mathematical modeling of poly[(R)-3-hydroxyalkanoate] synthesis by Cupriavidus necator DSM 545 on substrates stemming from biodiesel production. Bioresour Technol 133:482–494
Wang ZX, Bramer C, Steinbuchel A (2003) Two phenotypically compensating isocitrate dehydrogenases in Ralstonia eutropha. FEMS Microbiol Lett 227:9–16
Yu J, Si Y (2004) Metabolic carbon fluxes and biosynthesis of polyhydroxyalkanoates in Ralstonia eutropha on short chain fatty acids. Biotechnol Prog 20:1015–1024
Acknowledgments
This work was financially supported by the Collaborative EU-FP7 project ANIMPOL (“Biotechnological conversion of carbon containing wastes for eco-efficient production of high added value products”; Grant agreement No.: 245084). We are thankful to Dr. Matthias Raberg and Prof. Alexander Steinbüchel (UNI Münster), Dr. Anja Pöhlein and Prof. Rolf Daniel (UNI Göttingen) for useful information provided.
Conflict of interest
All authors have agreed to submit the manuscript to the Journal of Industrial Microbiology and Biotechnology. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2014_1439_MOESM1_ESM.doc
Online resource 1: ESM 1.doc (Metabolic reactions, list of chemical reactions for established metabolic network). (DOC 51 kb)
10295_2014_1439_MOESM3_ESM.xls
Online resource 3: ESM 3.xls (Matrix A_NAD, matrix with stoichiometric coefficients of internal metabolites in case when NAD and NADH are reactants in reaction of GLY-3-P DH). (XLS 47 kb)
10295_2014_1439_MOESM4_ESM.xls
Online resource 4: ESM 4.xls (Matrix A_FAD, matrix with stoichiometric coefficients of internal metabolites in case when FAD and FADH are reactants in reaction of GLY-3-P DH). (XLS 45 kb)
Rights and permissions
About this article
Cite this article
Lopar, M., Špoljarić, I.V., Cepanec, N. et al. Study of metabolic network of Cupriavidus necator DSM 545 growing on glycerol by applying elementary flux modes and yield space analysis. J Ind Microbiol Biotechnol 41, 913–930 (2014). https://doi.org/10.1007/s10295-014-1439-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1439-y