Abstract
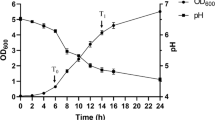
Lactobacillus casei Zhang is a widely recognized probiotic bacterium, which is being commercially used in China. To study the gene expression dynamics of L. casei Zhang during fermentation in soymilk, a whole genome microarray was used to screen for differentially expressed genes when grown to the lag phase, the late logarithmic phase, and the stationary phase. Comparisons of different transcripts next to each other revealed 162 and 63 significantly induced genes in the late logarithmic phase and stationary phase, of which the expression was at least threefold up-regulated and down-regulated, respectively. Approximately 38.4% of the up-regulated genes were associated with amino acid transport and metabolism notably for histidine and lysine biosynthesis, followed by genes/gene clusters involved in carbohydrate transport and metabolism, lipid transport and metabolism, and inorganic ion transport and metabolism. The analysis results suggest a complex stimulatory effect of soymilk-based ecosystem on the L. casei Zhang growth. On the other hand, it provides the very first insight into the molecular mechanism of L. casei strain for how it will adapt to the protein-rich environment.



Similar content being viewed by others
References
Azcarate-Peril MA, McAuliffe O, Altermann E, Lick S, Russell WM, Klaenhammer TR (2005) Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl Environ Microbiol 71:5794–5804
Boyaval P (1989) Lactic acid bacteria and metal ions. Lait 69:87–113
Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL (2009) Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 1:239–257
Cappa F, Cattivelli D, Cocconcelli PS (2005) The uvrA gene is involved in oxidative and acid stress responses in Lactobacillus helveticus CNBL1156. Res Microbiol 156:1039–1047
Chang YY, Cronan JE Jr (1999) Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol 33:249–259
Christensen JE, Dudley EG, Pederson JA, Steele JL (1999) Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 76:217–246
Corcoran BM, Stanton C, Fitzgerald G, Ross RP (2008) Life under stress: the probiotic stress response and how it may be manipulated. Curr Pharm Des 14:1382–1399
Cotter PD, Hill C (2003) Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453
Dave RI, Shah NP (1998) Ingredient supplementation effects on viability of probiotic bacteria in yogurt. J Dairy Sci 81:2804–2816
De Vuyst L (2000) Technology aspects related to the application of functional starter cultures. Food Technol Biotechnol 38:105–112
den Hengst CD, Groeneveld M, Kuipers OP, Kok J (2006) Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J Bacteriol 188:3280–3289
Donkor ON, Shah NP (2008) Production of beta-glucosidase and hydrolysis of isoflavone phytoestrogens by Lactobacillus acidophilus, Bifidobacterium lactis, and Lactobacillus casei in soymilk. J Food Sci 73:M15–M20
Duwat P, Ehrlich SD, Gruss A (1995) The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol Microbiol 17:1121–1131
Fozo EM, Kajfasz JK, Quivey RG Jr (2004) Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol Lett 238:291–295
Fozo EM, Quivey RG Jr (2004) The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J Bacteriol 186:4152–4158
Franck P, Moneret Vautrin DA, Dousset B, Kanny G, Nabet P, Guenard-Bilbaut L, Parisot L (2002) The allergenicity of soybean-based products is modified by food technologies. Int Arc Allergy Immunol 128:212–219
Gerharz T, Reinelt S, Kaspar S, Scapozza L, Bott M (2003) Identification of basic amino acid residues important for citrate binding by the periplasmic receptor domain of the sensor kinase CitA. Biochemistry 42:5917–5924
Gorke B, Stulke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624
Guedon E, Renault P, Ehrlich SD, Delorme C (2001) Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J Bacteriol 183:3614–3622
Guo J, Wang J, Yan L, Chen W, Liu X, Zhang H (2009) In vitro comparison of probiotic properties of Lactobacillus casei Zhang, a potential new probiotic, with selected probiotic strains. Lebenson Wiss Technol 42:1640–1646
Haandrikman A, Kok JJ, Venema G (1991) Lactococcal proteinase maturation protein PrtM is a lipoprotein. J Bacteriol 173:4517–4525
Juillard V, Le Bars D, Kunji ER, Konings WN, Gripon JC, Richard J (1995) Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl Environ Microbiol 61:3024–3030
Kunji ER, Hagting A, De Vries CJ, Juillard V, Haandrikman AJ, Poolman B, Konings WN (1995) Transport of beta-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis. J Biol Chem 270:1569–1574
Laddaga RA, Bessen R, Silver S (1985) Cadmium-resistant mutant of Bacillus subtilis 168 with reduced cadmium transport. J Bacteriol 162:1106–1110
Li H, Lu M, Guo H, Li W, Zhang H (2010) Protective effect of sucrose on the membrane properties of Lactobacillus casei Zhang subjected to freeze-drying. J Food Protect 73:715–719
Lin FM, Chiu CH, Pan TM (2004) Fermentation of a milk-soymilk and Lycium chinense Miller mixture using a new isolate of Lactobacillus paracasei subsp. paracasei NTU101 and Bifidobacterium longum. J Ind Microbiol Biotechnol 31:559–564
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Luesink EJ, van Herpen RE, Grossiord BP, Kuipers OP, de Vos WM (1998) Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol 30:789–798
Matsubara K, Ohnishi K, Kiritani K (1992) Nucleotide sequences and characterization of liv genes encoding components of the high-affinity branched-chain amino acid transport system in Salmonella typhimurium. J Biochem 112:93–101
Nies DH, Silver S (1995) Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol 14:186–199
O’Connell-Motherway M, van Sinderen D, Morel-Deville F, Fitzgerald GF, Ehrlich SD, Morel P (2000) Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935–947
Pastar I, Tonic I, Golic N, Kojic M, van Kranenburg R, Kleerebezem M, Topisirovic L, Jovanovic G (2003) Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl Environ Microbiol 69:5802–5811
Poolman B, Molenaar D, Smid EJ, Ubbink T, Abee T, Renault PP, Konings WN (1991) Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J Bacteriol 173:6030–6037
Quattrucci E, Bruschi L, Manzi P, Aromolo R, Panfili G (1997) Nutritional evaluation of typical and reformulated Italian cheeses. J Sci Food Agr 73:46–52
Quay SC, Dick TE, Oxender DL (1977) Role of transport systems in amino acid metabolism: leucine toxicity and the branched-chain amino acid transport systems. J Bacteriol 129:1257–1265
Quivey RG Jr, Faustoferri R, Monahan K, Marquis R (2000) Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans. FEMS Microbiol Lett 189:89–92
Redon E, Loubiere P, Cocaign-Bousquet M (2005) Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol 187:3589–3592
Reid G (2005) The importance of guidelines in the development and application of probiotics. Curr Pharm Des 11:11–16
Renault P, Gaillardin C, Heslot H (1988) Role of malolactic fermentation in lactic acid bacteria. Biochimie 70:375–379
Rico J, Yebra MJ, Perez-Martinez G, Deutscher J, Monedero V (2008) Analysis of ldh genes in Lactobacillus casei BL23: role on lactic acid production. J Ind Microbiol Biotechnol 35:579–586
Saulnier D, Molenaar MD, WMd Vos, Gibson GR, Kolida S (2007) Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microbiol 73:1753–1765
Savijoki K, Ingmer H, Varmanen P (2006) Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol 71:394–406
Shah NP (2000) Probiotic bacteria: selective enumeration and survival in dairy foods. J Dairy Sci 83:894–907
Sheng J, Marquis RE (2007) Malolactic fermentation by Streptococcus mutans. FEMS Microbiol Lett 272:196–201
Shimakawa Y, Matsubara S, Yuki N, Ikeda M, Ishikawa F (2003) Evaluation of Bifidobacterium breve strain Yakult-fermented soymilk as a probiotic food. Int J Food Microbiol 81:131–136
Singh VK, Moskovitz J (2003) Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149:2739–2747
Siro I, Kapolna E, Kapolna B, Lugasi A (2008) Functional food. Product development, marketing and consumer acceptance–a review. Appetite 51:456–467
Stucky K, Hagting A, Klein JR, Matern H, Henrich B, Konings WN, Plapp R (1995) Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbruckii subsp. lactis DSM7290. Mol Gen Genet 249:682–690
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278:631–637
Trindade CS, Terzi SC, Trugo LC, Della Modesta RC, Couri S (2001) Development and sensory evaluation of soy milk based yoghurt. Arch Latinoamericanos Nutr 51:100–104
Tynecka Z, Gos Z, Zajac J (1981) Reduced cadmium transport determined by a resistance plasmid in Staphylococcus aureus. J Bacteriol 147:305–312
van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, Maguin E (2002) Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216
Wall T, Bath K, Britton RA, Jonsson H, Versalovic J, Roos S (2007) The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl Environ Microbiol 73:3924–3935
Wang H, Dong C, Chen Y, Cui L, Zhang H (2010) A new probiotic cheddar cheese with high ACE-inhibitory activity and γ-Aminobutyric acid content produced with koumiss-derived Lactobacillus casei Zhang. Food Technol Biotechnol 48:62–70
Wang J, Guo Z, Zhang Q, Yan L, Chen W, Liu XM, Zhang HP (2009) Fermentation characteristics and transit tolerance of probiotic Lactobacillus casei Zhang in soymilk and bovine milk during storage. J Dairy Sci 92:2468–2476
Wu R, Wang L, Wang J, Li H, Menghe B, Wu J, Guo M, Zhang H (2009) Isolation and preliminary probiotic selection of lactobacilli from koumiss in Inner Mongolia. J Basic Microbiol 49:318–326
Wu R, Wang W, Yu D, Zhang W, Li Y, Sun Z, Wu J, Meng H, Zhang H (2009) Proteomic analysis of Lactobacillus casei Zhang, a new probiotic bacterium isolated from traditionally home-made koumiss in Inner Mongolia of China. Mol Cell Proteomics 8:2321–2338
Ya T, Zhang Q, Chu F, Merritt J, Bilige M, Sun T, Du R, Zhang H (2008) Immunological evaluation of Lactobacillus casei Zhang: a newly isolated strain from koumiss in Inner Mongolia, China. BMC Immunol 9:68
Yan D (2007) Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci USA 104:9475–9480
Zhang H, Xu J, Wang J, Menghebilige SunT, Li H, Guo M (2008) A survey on chemical and microbiological composition of kurut, naturally fermented yak milk from Qinghai in China. Food Control 19:578–586
Zhang W, Yu D, Sun Z, Chen X, Bao Q, Meng H, Hu S, Zhang H (2008) Complete nucleotide sequence of plasmid plca36 isolated from Lactobacillus casei Zhang. Plasmid 60:131–135
Zhang W, Yu D, Sun Z, Wu R, Chen X, Chen W, Meng H, Hu S, Zhang H (2010) Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J Bacteriol 192:5268–5269
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 30860219), the Hi-tech Research and Development Program of China (863 Program) (Grant No. 2010AA10Z302), the earmarked fund for Modern Agro-industry Technology Research System (Grant No. nycytx-0501), and the Innovation Research Team Development Program of Ministry of Education of China (Grant No. IRT0967). We would like to thank Eric Richard Spaans for the English editing.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ji-Cheng Wang and Wen-Yi Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, JC., Zhang, WY., Zhong, Z. et al. Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk. J Ind Microbiol Biotechnol 39, 191–206 (2012). https://doi.org/10.1007/s10295-011-1015-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-1015-7