Abstract
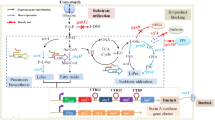
β-1,6-N-Acetylglucosaminidase (dispersin B), which cleaves poly-ß-(1,6)-linked N-acetylglucosamine, is encoded by dspB of Aggregatibacter actinomycetemcomitans. To enhance the production of dispersin B, we engineered dspB to transcribe mRNAs devoid of the trinucleotide ACA. Transcription and translation levels of ACA-less and wild-type dspB expressed in Escherichia coli (E. coli) under T5 and T7 promoters were analyzed by real-time RT-PCR and protein quantification, respectively. The ACA-less dspB mRNA level was significantly higher (P < 0.01) and produced 77.6 and 34.9% more dispersin B than wild-type dspB expressed under T7 and T5 promoters, respectively. Dispersin B expression under T7 promoter caused a 98–99.5% drop in the glyceraldehyde-3-phosphate dehydrogenase (gapA) mRNA level, which was not observed with T5 promoter. Fusion of green fluorescent protein (GFP) with dispersin B allowed rapid quantification of dispersin B production by measuring fluorescence intensity in culture broth. Although the cultures containing 0.1% glucose showed sustained increase in dispersin B-GFP production until 12 h, no significant increase in dispersin B activity was observed beyond 4 and 6 h after induction when expressed under T7 and T5 promoters, respectively. This study demonstrates the effectiveness of ACA-less mRNA and the advantage of GFP tagging for enhanced dispersin B production and quantification, which could be adapted for improving the production of other commercially important proteins in E. coli.




Similar content being viewed by others
References
Aizenman E, Engelberg-Kulka H, Glaser G (1996) An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA 93:6059–6063
Böhm D, Rinas U (2006) Comparative transcriptional profiling of the bacterial stress response in temperature and chemically-induced recombinant E. coli processes. Microb Cell Fact 5:1
Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38
Cserjan-Puschmann M, Kramer W, Duerrschmid E, Striedner G, Bayer K (1999) Metabolic approaches for the optimization of recombinant fermentation processes. Appl Microbiol Biotechnol 53:43–50
Dedhia N, Richins R, Mesina A, Chen W (1997) Improvement in recombinant protein production in ppGpp-deficient Escherichia coli. Biotechnol Bioeng 53:379–386
Hoffmann F, Rinas U (2004) Stress induced by recombinant protein production by Escherichia coli. Adv Biochem Eng Biotechnol 89:73–92
Itoh Y, Wang X, Hinnebusch BJ, Preston JF, Romeo T (2005) Depolymerization of ß-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol 187:382–387
Izano EA, Sadovskaya I, Vinogradov E, Mulks MH, Velliyagounder K, Ragunath C, Kher WB, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB (2007) Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog 43:1–9
Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T (2002) Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol 184:290–301
Jones JJ, Bridges AM, Fosberry AP, Gardner S, Lowers RR, Newby RR, James PJ, Hall RM, Jenkins O (2004) Potential of real-time measurement of GFP-fusion proteins. J Biotechnol 109:201–211
Kaplan JB, Ragunath C, Ramasubbu N, Fine DH (2003) Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous ß-hexosaminidase activity. J Bacteriol 185:4693–4698
Kolodkin-Gal I, Engelberg-Kulka H (2006) Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J Bacteriol 188:3420–3423
Omoya K, Kato Z, Matsukuma E, Li A, Hashimoto K, Yamamoto Y, Ohnishi H, Kondo N (2004) Systematic optimization of active protein expression using GFP as a folding reporter. Protein Expr Purif 36:327–332
Rücker E, Schneider G, Steinhäuser K, Löwer R, Hauber J, Stauber RH (2001) Rapid evaluation and optimization of recombinant protein production using GFP tagging. Protein Expr Purif 21:220–223
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H (2001) Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol 183:2041–2045
Smith PK, Krohn RI, Hermanson GT, Malli AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
Suzuki M, Roy R, Zheng H, Woychik N, Inouye M (2006) Bacterial bioreactors for high yield production of recombinant protein. J Biol Chem 281:37559–37565
Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M (2005) Single protein production in living cells facilitated by an mRNA interferase. Mol Cell 18:253–261
Wang X, Preston JF, Romeo T (2004) The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186:2724–2734
Yakandawala N, Romeo T, Friesen AD, Madhyatha S (2008) Metabolic engineering of Escherichia coli to enhance phenylalanine production. Appl Microbiol Biotechnol 78:283–291
Zhang Y, Zhang J, Hara H, Kato I, Inouye M (2004) Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem 280:3143–3150
Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M (2003) MazEF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12:913–923
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yakandawala, N., Gawande, P.V., LoVetri, K. et al. Enhanced expression of engineered ACA-less β-1, 6-N-acetylglucosaminidase (dispersin B) in Escherichia coli . J Ind Microbiol Biotechnol 36, 1297–1305 (2009). https://doi.org/10.1007/s10295-009-0613-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0613-0