Abstract
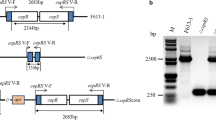
Three open reading frames denoted as orf21, orf22, and orf23 were identified from downstream of the currently recognized gene cluster for clavulanic acid biosynthesis in Streptomyces clavuligerus ATCC 27064. The new orfs were annotated after in silico analysis as genes encoding a putative sigma factor, a sensor kinase, and a response regulator. The roles of the individual genes were explored by disruption of the corresponding orfs, and the morphological and antibiotic production phenotypes of the resulting mutants were compared. In orf21 and orf22 mutants, no growth or morphological differences were noted, but modest reduction of cephamycin C (orf21), or both cephamycin C and clavulanic acid production (orf22) compared with wild-type, were observed. In orf23 mutant, cell growth and sporulation was retarded, and clavulanic acid and cephamycin C production were reduced to 40 and 47% of wild-type levels, respectively. Conversely, overexpression of orf23 caused precocious hyperproduction of spores on solid medium, and antibiotic production was increased above the levels seen in plasmid control cultures. Transcriptional analyses were also carried out on orf23 and showed that mutation had little effect on transcription of genes associated with the early stages of cephamycin C or clavulanic acid production but transcription of claR, which regulates the late stages of clavulanic acid production, was reduced in orf23 mutants. These observations suggest that the orf23 product may enable S. clavuligerus to respond to environmental changes by altering cell growth and differentiation. In addition, the effects of ORF23 on growth might indirectly regulate the biosynthesis of secondary metabolites such as clavulanic acid and cephamycin C.





Similar content being viewed by others
References
Alexander D, Jensen SE (1998) Investigation of the Streptomyces clavuligerus cephamycin gene cluster and its regulation by the CcaR. J Bacteriol 180:4068–4079
Aravind L, Ponting CP (1999) The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signaling proteins. FEMS Microbiol Lett 176:111–116. doi:10.1016/S0378-1097(99)00197-4; doi:10.1111/j.1574-6968.1999.tb13650.x
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi:10.1038/417141a
Brian P, Riggle PJ, Santos RA, Champness WC (1996) Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J Bacteriol 178:3221–3231
Brown AG, Butterworth D, Cole M, Hanscomb G, Hood JD, Reading C, Rolinson GN (1976) Naturally occurring β-lactamase inhibitors with antibacterial activity. J Antibiot 29:668–669
Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA (2002) Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell 9:527–539. doi:10.1016/S1097-2765(02)00470-7
Chang HM, Chen MY, Shieh YT, Bibb MJ, Chen CW (1996) The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol Microbiol 21:1075–1085
Chater KF, Bibb MJ (1997) Regulation of bacterial antibiotic production. In: Kleinkauf H, von Dohren H (eds) Products of secondary metabolism, biotechnology, vol 6. VCH, Weinheim, pp 57–105
Dutta R, Inouye M (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25:24–28. doi:10.1016/S0968-0004(99)01503-0
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546. doi:10.1073/pnas.0337542100
Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ (2004) Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150:2795–2806. doi:10.1099/mic.0.27181-0
Ishizuka H, Horinouchi S, Kieser HM, Hopwood DA, Beppu T (1992) A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol 23:7585–7594
Jensen SE, Elder KJ, Aidoo KA, Paradkar AS (2000) Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob Agents Chemother 44:720–726. doi:10.1128/AAC.44.3.720-726.2000
Jensen SE, Paradkar AS, Mosher RH, Anders C, Beatty PH, Brumlik MJ, Griffin A, Barton B (2004) Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother 48:192–202. doi:10.1128/AAC.48.1.192-202.2004
Jin W, Kim HK, Kim JY, Kang SG, Lee SH, Lee KJ (2004) Cephamycin C production is regulated by relA and rsh genes in Streptomyces clavuligerus ATCC27064. J Biotechnol 114:81–87. doi:10.1016/j.jbiotec.2004.06.010
Kenney LJ (2002) Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5:135–141. doi:10.1016/S1369-5274(02)00310-7
Khaleeli N, Li R, Townsend CA (1999) Origin of the β-lactam carbons in clavulanic acid from an unusual thiamine pyrophosphate-mediated reaction. J Am Chem Soc 121:9223–9224. doi:10.1021/ja9923134
Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Li R, Khaleeli N, Townsend CA (2000) Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Bacteriol 182:4087–4095. doi:10.1128/JB.182.14.4087-4095.2000
Malmberg LH, Hu WS, Sherman DH (1993) Precusor flux control through targeted chromosomal insertion of the lysine ε-aminotransferase (lat) gene in cephamycin C biosynthesis. J Bacteriol 175:6916–6924
Mellado E, Lorenzana LM, Rodríguez-Sáiz M, Díez B, Liras P, Barredo JL (2002) The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 148:1427–1438
Netolitzky DJ, Wu X, Jensen SE, Roy KL (1995) Giant linear plasmids of β-lactam antibiotic producing Streptomyces. FEMS Microbiol Lett 131:27–34. doi:10.1016/0378-1097(95)00230-3
Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU (1998) In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol 143:901–910. doi:10.1083/jcb.143.4.901
Paradkar AS, Aidoo KA, Jensen SE (1998) A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol 27:831–843. doi:10.1046/j.1365-2958.1998.00731.x
Paradkar AS, Mosher RH, Anders C, Griffin A, Griffin J, Hughes C, Greaves P, Barton B, Jensen SE (2001) Applications of gene replacement technology to Streptomyces clavuligerus strain development for clavulanic acid production. Appl Environ Microbiol 67:2292–2297. doi:10.1128/AEM.67.5.2292-2297.2001
Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF (1997) A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol 179:2053–2059
Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P (1998) The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211:311–321. doi:10.1016/S0378-1119(98)00106-1
Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P (1999) Deletion of the pyc gene blocks clavulanic acid biosynthesis except in glycerol-containing medium: evidence for two different genes in formation of the C3 unit. J Bacteriol 181:6922–6928
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sola-Landa A, Moura RS, Martín JF (2003) The two component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci USA 100:6133–6138. doi:10.1073/pnas.0931429100
Solá M, Gomis-Rüth FX, Serrano L, González A, Coll M (1999) Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol 285:675–687. doi:10.1006/jmbi.1998.2326
Song JY, Kim ES, Kim DW, Jensen SE, Lee KJ (2008) Functional effects of increased copy number of the gene encoding proclavaminate amidino hydrolase on clavulanic acid production in Streptomyces clavuligerus ATCC 27064. J Microbiol Biotechnol 3:417–426
Tahlan K, Park HU, Jensen SE (2004) Three unlinked gene clusters are involved in clavam metabolite biosynthesis in Streptomyces clavuligerus. Can J Microbiol 50:803–810. doi:10.1139/w04-070
Tahlan K, Park HU, Wong A, Beatty PH, Jensen SE (2004) Two sets of paralogous genes encode the enzymes involved in the early stages of clavulanic acid and clavam biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother 48:930–939. doi:10.1128/AAC.48.3.930-939.2004
Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, Qian H, Inouye M, Ikura M (1999) Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol 6:729–734. doi:10.1038/11495
Ward JM, Hodgson JE (1993) The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘supercluster’ in three Streptomyces. FEMS Microbiol Lett 110:239–242. doi:10.1111/j.1574-6968.1993.tb06326.x
Acknowledgments
This work was supported by the KICOS through a grant provided by the Korean Ministry of Science & Technology (MOST) in 2007 (K20726000001-07B0100-00110), for collaborative research with the ActinoGEN project (IP005224) in the EU. The authors J. Y. Song, E. S. Kim, and D. W. Kim were supported by the second stage of the Brain Korea 21 project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, J.Y., Kim, E.S., Kim, D.W. et al. A gene located downstream of the clavulanic acid gene cluster in Streptomyces clavuligerus ATCC 27064 encodes a putative response regulator that affects clavulanic acid production. J Ind Microbiol Biotechnol 36, 301–311 (2009). https://doi.org/10.1007/s10295-008-0499-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0499-2