Abstract
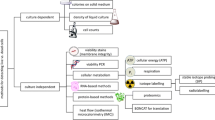
The majority of microorganisms have yet to be cultivated and represent a vast uncharacterized and untapped resource. Here, we report the utilization of a combination of flow cytometry, cultivation, and molecular genetics to develop new methodologies to access and characterize biodiversity in microbial samples. We demonstrate that fluorescent dyes and combinations of dyes can selectively stain portions of bacterial populations that can be isolated as sub-populations using fluorescence-activated cell sorting (FACS). Microbial sub-populations obtained by FACS differ substantially from the original microbial population, as demonstrated by denaturing gradient gel electrophoresis and determination of 16S rRNA gene sequences. These sub-populations can subsequently be used to inoculate microbial growth media, allowing the isolation of different microbial species from those that can be readily cultivated from the original sample using the same microbial growth media. When this technique was applied to the analysis of activated-sludge and Yellowstone Lake hydrothermal vent samples, comparative analysis of 16S rDNA sequences revealed that FACS allowed the detection of numerous bacterial species, including previously unknown species, not readily detectable in the original sample due to low relative abundance. This approach may result in a convenient methodology to more thoroughly characterize microbial biodiversity.



Similar content being viewed by others
References
Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF (2004) Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551–554
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amor KB, Breeuwer P, Verbaarschot P, Rombouts FM, Akkermans AD, De Vos WM, Abee T (2002) Multiparametric flow cytometry and cell sorting for the assessment of viable, injured, and dead bifidobacterium cells during bile salt stress. Appl Environ Microbiol 68:5209–5216
Barns SM, Fundyga RE, Jeffries MW, Pace NR (1994) Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA 91:1609–1613
Brosius J, Dull TJ, Sleeter DD, Noller HF (1981) Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol 148:107–127
Button DK, Robertson BR, Juttner F (1996) Microflora of a subalpine lake: bacterial populations, size and DNA distributions, and their dependence on phosphate. FEMS Microbiol Ecol 21:87–101
Button DK, Robertson BR, Lepp PW, Schmidt TM (1998) A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl Environ Microbiol 64:4467–4476
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol 35:1–21
Hewitt CJ, Nebe-Von-Caron G (2004) The application of multi-parameter flow cytometry to monitor individual microbial cell physiological state. Adv Biochem Eng Biotechnol 89:197–223
Holben WE, Feris KP, Kettunen A, Apajalahti JH (2004) GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:2263–2270
Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Baumler AJ (2003) The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype typhimurium fimbrial operons. Mol Microbiol 48:1357–1376
Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH (2003) Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol 69:7210–7215
Keller M, Zengler K (2004) Tapping into microbial diversity. Nat Rev Microbiol 2:141–150
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodman M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–173
Liesack W, Janssen PH, Rainey F, Ward-Rainey NL, Stackebrandt E (1997) Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. Mod Soil Microbiol 375–439
Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR Jr, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
Mason DJ, Shanmuganathan S, Mortimer FC, Gant VA (1998) A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl Environ Microbiol 64:2681–2685
Muller S, Strauber H, Losche A, Babel W (2002) Population analysis of a binary bacterial culture by multi-parametric flow cytometry. J Biotechnol 97:163–176
Muyzer G, Waal EC de, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nogales B, Moore ER, Llobet-Brossa E, Rossello-Mora R, Amann R, Timmis KN (2001) Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl Environ Microbiol 67:1874–1884
Ovreas L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Prigione V, Lingua G, Marchisio VF (2004) Development and use of flow cytometry for detection of airborne fungi. Appl Environ Microbiol 70:1360–1365
Raskin L, Zheng D, Griffin ME, Stroot PG, Misra P (1995) Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie van Leeuwenhoek 68:297–308
Resina-Pelfort O, Comas-Riu J, Vives-Rego J (2001) Effects of deflected droplet electrostatic cell sorting on the viability and exoproteolytic activity of bacterial cultures and marine bacterioplankton. Syst Appl Microbiol 24:31–36
Reysenbach AL, Wickham GS, Pace NR (1994) Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol 60:2113–2119
Schmalenberger A, Schwieger F, Tebbe CC (2001) Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl Environ Microbiol 67:3557–3563
Spiro A, Lowe M, Brown D (2000) A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl Environ Microbiol 66:4258–4265
Steen HB (1990) Light scattering measurement in an arc lamp-based flow cytometer. Cytometry 11:223–230
Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA (2004) New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol 70:4748–4755
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072
Tanaka Y, Yamaguchi N, Nasu M (2000) Viability of Escherichia coli O157:H7 in natural river water determined by the use of flow cytometry. J Appl Microbiol 88:228–236
Torsvik V, Goksoyr J, Daae FL (1990) High diversity in DNA of soil bacteria. Appl Environ Microbiol 56:782–787
Wallner G, Fuchs B, Spring S, Beisker W, Amann R (1997) Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol 63:4223–4231
Zoetendal EG, Ben-Amor K, Harmsen HJ, Schut F, Akkermans AD, Vos WM de (2002) Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl Environ Microbiol 68:4225–4232
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, HS., Schumacher, R. & Kilbane, J.J. New method to characterize microbial diversity using flow cytometry. J IND MICROBIOL BIOTECHNOL 32, 94–102 (2005). https://doi.org/10.1007/s10295-005-0208-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-005-0208-3