Abstract
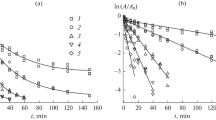
The autolysis of yeast cells has practical implications in the production of fermented foods and beverages and flavourants for food processing. Protein and RNA degradation during yeast autolysis are well described but the fate of DNA is unclear. Yeast cells (Saccharomyces cerevisiae) were autolysed by incubating suspensions at 30–60°C (pH 7.0), and at pH 4.0–7.0 (40°C) for 10–14 days. Up to 55% of total DNA was degraded, with consequent leakage into the extracellular environment of mainly 3′- and 5′-deoxyribonucleotides, and lesser amounts of polynucleotides. The rate and extent of DNA degradation, composition of the DNA degradation products and DNase activity were affected by temperature and pH. The highest amount of DNA degradation occurred at 40°C and pH 7.0, where the highest DNase activity was recorded. DNase activity was lowest at 60°C and pH 4.0, where the proportion of polynucleotides in the degradation products was higher.





Similar content being viewed by others
References
Adams RLP, Knowler JT, Leader DP (1993) The biochemistry of the nucleic acids, 11th edn. Chapman and Hall, London
Arnold WN (1981) Autolysis. In: Arnold WN (ed) Yeast cell envelopes: biochemistry, biophysics and ultrastructure. CRC Press, New York, pp 129–137
Babayan TL, Bezrukovv MG (1985) Autolysis of yeasts. Acta Biotechnol 5:129–136
Bryant DW, Haynes RHA (1984) DNA endonuclease isolated from yeast nuclease extract. Can J Biochem 56:181–189
Charpentier C, Feuillat M (1993) Yeast autolysis. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood, Chur, Switzerland, pp 225–242
Connew SJ (1998) Yeast autolysis. A review of current research. Aust N Z Wine Ind J 13:61–64
Dziezak JD (1987) Yeast and yeast derivatives: definitions, characteristics, and processing. Food Technol 41:104–125
Ezekiel UR, Zassenhaus HP (1993) Localization of a cruciform cutting endonuclease in yeast mitochondria. Mol Gen Genet 240:414–418
Farrer KTH (1956) The autolysis in yeasts. Food Sci Abstr 28:1–12
Gimble FS, Thorner J (1993) Purification and characterization of VDE, a site-specific endonuclease from the yeast Saccharomyces cerevisiae. J Biol Chem 268:21844–21853
Habraken Y, Sung P, Prakash L, Prakash S (1993) Yeast excision repair gene RAD2 encodes a single stranded DNA endonuclease. Nature 366:365–368
Halasz A, Lásztity R (1991) Use of yeast biomass in food production. CRC Press, Boca Raton, Fla.
Hernawan T, Fleet GH (1995) Chemical and cytological changes during the autolysis of yeasts. J Ind Microbiol 14:440–450
Hough MD, Maddox IS (1970) Yeast autolysis. Process Biochem 5:50–52
Joslyn MA (1955) Yeast autolysis. Chemical and cytological changes involved in autolysis. Wallerstein Lab Commun 18:107–119
Joslyn MA, Vosti DC (1955) Yeast autolysis. Factors influencing the rate and extent of autolysis. Wallerstein Lab Commun 18:191–210
Kinsella JE (1986) Functional proteins from yeast nucleoprotein for food uses: method for isolation. In: Knorr D, Dekker M (eds) Food biotechnology. Dekker, New York, pp 363–391
Labarca C, Paigen K (1980) A simple, rapid and sensitive DNA assay procedure. Anal Biochem 102:344–352
Martinez-Rodriguez AJ, Polo MC (2000) Characterization of the nitrogen compounds released during yeast autolysis in a model wine system. J Agric Food Chem 48:1081–1085
Martinez-Rodriguez AJ, Carrascosa AV, Polo MC (2001) Release of nitrogen compounds to the extracellular medium by three strains of Saccharomyces cerevisiae during induced autolysis in a model wine system. Int J Food Microbiol 64:155–160
Masschelein CA (1986) Centenary review: the biochemistry of maturation. J Inst Brew 92:213–219
Morosoli R, Lusena CV (1980) An endonuclease from yeast mitochondrial fractions. Eur J Biochem 110:431–437
Nagodawithana T (1992) Yeast-derived flavors and flavor enhancers and their probable mode of action. Food Technol 46:138–140
Nagodawithana T (1993) Enzymes associated with savory flavor enhancement. In: Nagodawithana T, Reed G (eds) Enzymes in food processing, 3rd edn. Academic Press, New York, pp 401–421
Piñon R, Leney E (1975). Studies on deoxyribonucleases from Saccharomyces cerevisiae. Characterization of two endonuclease activities with a preference for double-stranded DNA. Nucleic Acids Res 2:1023–1042
Schapira M, Desdouets C, Jacq C, Perea J (1993) I-Sec III an intron-encoded DNA endonuclease from yeast mitochondria. Nucleic Acids Res 21:3683–3689
Stam H, Hoogland M, Laane C (1998) Food flavours from yeast. In: Wood BJ (ed) Microbiology of fermented foods, vol 2, 2nd edn. Blackie, London, pp 505–542
Sung P, Reynolds P, Prakash L, Prakash S (1993) Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J Biol Chem 268:26391–26399
Suomalainen H (1975) Some enzymological factors influencing the leavening capacity and keeping quality of baker's yeast. Eur J Appl Microbiol 1:1–12
Takakuwa M, Watanabe Y (1981) Degradation of cellular phospholipids and softening of pressed baker's yeast. Agric Biol Chem 45:2167–2173
Todd BE, Fleet GH, Henschke PA (2000) Promotion of autolysis through the interaction of killer and sensitive yeasts: potential application in sparkling wine production. Am J Enol Vitic 51:65–72
Trevelyan WE (1977) Induction of autolytic breakdown of RNA in yeast by addition of ethanol and by drying/rehydration. J Sci Food Agric 28:579–588
Trevelyan WE (1978) Effect of procedures for the reduction of the nucleic acid content of SCP on the DNA content of Saccharomyces cerevisiae. J Sci Food Agric 29:903–908
Williamson DH (1991) Nucleus, chromosomes and plasmids. In: Rose AH, Harrison JS (eds) The yeasts, vol 4. Yeast organelles, 2nd edn. Academic Press, London, pp 433–488
Zhao J, Fleet GH (1996) Separation of 20 isomers of ribonucleotides and deoxyribonucleotides by reversed-phase ion-pairing high-performance liquid chromotography. J Chromotogr A 732:271–275
Zhao J, Todd BE, Fleet GH (1994) Separation of ribonucleotides, ribonucleosides, deoxyribonucleotides, deoxyribonucleosides and bases by reversed-phase high-performance liquid chromotography. J Chromotogr A 673:167–171
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, J., Fleet, G.H. Degradation of DNA during the autolysis of Saccharomyces cerevisiae . J IND MICROBIOL BIOTECHNOL 30, 175–182 (2003). https://doi.org/10.1007/s10295-003-0028-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-003-0028-2