Abstract
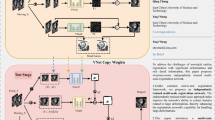
Image registration is a fundamental task in various applications of medical image analysis and plays a crucial role in auxiliary diagnosis, treatment, and surgical navigation. However, cardiac image registration is challenging due to the large non-rigid deformation of the heart and the complex anatomical structure. To address this challenge, this paper proposes an independently trained multi-scale registration network based on an image pyramid. By down-sampling the original input image multiple times, we can construct image pyramid pairs, and design a multi-scale registration network using image pyramid pairs of different resolutions as the training set. Using image pairs of different resolutions, train each registration network independently to extract image features from the image pairs at different resolutions. During the testing stage, the large deformation registration is decomposed into a multi-scale registration process. The deformation fields of different resolutions are fused by a step-by-step deformation method, thereby addressing the challenge of directly handling large deformations. Experiments were conducted on the open cardiac dataset ACDC (Automated Cardiac Diagnosis Challenge); the proposed method achieved an average Dice score of 0.828 in the experimental results. Through comparative experiments, it has been demonstrated that the proposed method effectively addressed the challenge of heart image registration and achieved superior registration results for cardiac images.




Similar content being viewed by others
Data Availability
The data that support the findings of this study are openly available in “Training dataset” at https://www.creatis.insa-lyon.fr/Challenge/acdc/databasesTraining.html.
References
Khalil A, Ng S C, Liew Y M, et al. An overview on image registration techniques for cardiac diagnosis and treatment[J]. Cardiology research and practice, 2018, 2018.
Klein G J, Huesman R H. Four-dimensional processing of deformable cardiac PET data[J]. Medical Image Analysis, 2002, 6(1): 29-46.
Rogers Jr W J, Shapiro E P, Weiss J L, et al. Quantification of and correction for left ventricular systolic long-axis shortening by magnetic resonance tissue tagging and slice isolation[J]. Circulation, 1991, 84(2): 721-731.
O'Dell W G, Moore C C, Hunter W C, et al. Three-dimensional myocardial deformations: calculation with displacement field fitting to tagged MR images[J]. Radiology, 1995, 195(3): 829-835.
Avendi M R, Kheradvar A, Jafarkhani H. Automatic segmentation of the right ventricle from cardiac MRI using a learning‐based approach[J]. Magnetic resonance in medicine, 2017, 78(6): 2439-2448.
Tavakoli V, Amini A A. A survey of shaped-based registration and segmentation techniques for cardiac images[J]. Computer Vision and Image Understanding, 2013, 117(9): 966-989.
Zhou Y, Pang S, Cheng J, et al. Unsupervised deformable medical image registration via pyramidal residual deformation fields estimation[J]. arXiv preprint arXiv:2004.07624, 2020.
X Hu M Kang W Huang Dual-stream pyramid registration network[C], , Medical Image Computing and Computer Assisted Intervention–MICCAI, et al 2019 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings Part II. Cham: Springer International Publishing 2019 382 390
Zhang L, Zhou L, Li R, et al. Cascaded feature warping network for unsupervised medical image registration[C]//2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). IEEE, 2021: 913–916.
Dosovitskiy A, Fischer P, Ilg E, et al. Flownet: Learning optical flow with convolutional networks[C]//Proceedings of the IEEE international conference on computer vision. 2015: 2758–2766.
Cao Y, Zhu Z, Rao Y, et al. Edge-aware pyramidal deformable network for unsupervised registration of brain MR images[J]. Frontiers in Neuroscience, 2021, 14: 620235.
Fechter, Tobias, Dimos Baltas. One-shot learning for deformable medical image registration and periodic motion tracking. IEEE transactions on medical imaging 39.7 (2020): 2506-2517.
Yu, Hanchao, et al. Motion pyramid networks for accurate and efficient cardiac motion estimation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part VI 23. Springer International Publishing, 2020.
Li H, Fan Y, Alzheimer's Disease Neuroimaging Initiative. MDReg‐Net: Multi‐resolution diffeomorphic image registration using fully convolutional networks with deep self‐supervision[J]. Human Brain Mapping, 2022, 43(7): 2218–2231.
TCW Mok ACS Chung Large deformation diffeomorphic image registration with Laplacian pyramid networks[C], , Medical Image Computing and Computer Assisted Intervention–MICCAI, 2020 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part III 23 Springer International Publishing 2020 211 221
Jiang Z, Yin F F, Ge Y, et al. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration[J]. Physics in Medicine & Biology, 2020, 65(1): 015011.
Eppenhof K A J, Lafarge M W, Veta M, et al. Progressively trained convolutional neural networks for deformable image registration[J]. IEEE transactions on medical imaging, 2019, 39(5): 1594-1604.
Milletari F, Navab N, Ahmadi S A. V-net: Fully convolutional neural networks for volumetric medical image segmentation[C]//2016 fourth international conference on 3D vision (3DV). Ieee, 2016: 565–571.
Bernard O, Lalande A, Zotti C, et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved?[J]. IEEE transactions on medical imaging, 2018, 37(11): 2514-2525.
Ketkar N. Introduction to pytorch[J]. Deep Learning with Python, Springer, 2018: 195-208.
Kingma P, Ba J. Adam: A method for stochastic optimization[J]. arXiv preprint arXiv:1412.6980, 2014.
Lee R. Dice. Measures of the amount of ecologic association between species[J]. Ecology, 1945, 26(3): 297-302.
Thirion J P. Image matching as a diffusion process: an analogy with Maxwell's demons[J]. Medical image analysis, 1998, 2(3): 243-260.
Avants B B, Epstein C L, Grossman M, et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain[J]. Medical image analysis, 2008, 12(1): 26-41.
Balakrishnan G, Zhao A, Sabuncu M R, et al. Voxelmorph: a learning framework for deformable medical image registration[J]. IEEE transactions on medical imaging, 2019, 38(8): 1788-1800.
Avants B B, Tustison N, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration [J]. NeuroImage, 2011, 54(3): 2033-2044.
Funding
This work was supported by the National Natural Science Foundation of China under award number 61976091.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
This is an observational study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, Q., Wang, Y. & Zhang, J. Independently Trained Multi-Scale Registration Network Based on Image Pyramid. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01019-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01019-8