Abstract
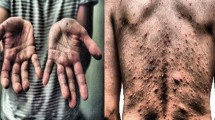
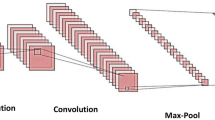
Monkeypox (MPox) is an infectious disease caused by the monkeypox virus, presenting challenges in accurate identification due to its resemblance to other diseases. This study introduces a deep learning-based method to distinguish visually similar diseases, specifically MPox, chickenpox, and measles, addressing the 2022 global MPox outbreak. A two-stage optimization approach was presented in the study. By analyzing pre-trained deep neural networks including 71 models, this study optimizes accuracy through transfer learning, fine-tuning, and ensemble learning techniques. ConvNeXtBase, Large, and XLarge models were identified achieving 97.5% accuracy in the first stage. Afterwards, some selection criteria were followed for the models identified in the first stage for use in ensemble learning technique within the optimization approach. The top-performing ensemble model, EM3 (composed of RegNetX160, ResNetRS101, and ResNet101), attains an AUC of 0.9971 in the second stage. Evaluation on unseen data ensures model robustness and enhances the study’s overall validity and reliability. The design and implementation of the study have been optimized to address the limitations identified in the literature. This approach offers a rapid and highly accurate decision support system for timely MPox diagnosis, reducing human error, manual processes, and enhancing clinic efficiency. It aids in early MPox detection, addresses diverse disease challenges, and informs imaging device software development. The study’s broad implications support global health efforts and showcase artificial intelligence potential in medical informatics for disease identification and diagnosis.









Similar content being viewed by others
Data Availability
The dataset used in this article is online available at https://doi.org/10.17632/R9BFPNVYXR.6.
References
WHO. (2023). Mpox (monkeypox). Mpox (Monkeypox). https://www.who.int/news-room/fact-sheets/detail/monkeypox
Haque, Md. E., Ahmed, Md. R., Nila, R. S., & Islam, S. (2022a). Classification of Human Monkeypox Disease Using Deep Learning Models and Attention Mechanisms. https://arxiv.org/abs/2211.15459v1
CDC. (2022). About Chickenpox. About Chickenpox. https://www.cdc.gov/chickenpox/about/index.html#
NHS. (2022). Measles. Measles. https://www.nhs.uk/conditions/measles/
Delidow, B. C., Lynch, J. P., Peluso, J. J., & White, B. A. (1993). Polymerase Chain Reaction. In B. A. White (Ed.), PCR Protocols: Current Methods and Applications (pp. 1–29). Humana Press. https://doi.org/10.1385/0-89603-244-2:1
Binny, R. N., Priest, P., French, N. P., Parry, M., Lustig, A., Hendy, S. C., Maclaren, O. J., Ridings, K. M., Steyn, N., Vattiato, G., & Plank, M. J. (2023). Sensitivity of Reverse Transcription Polymerase Chain Reaction Tests for Severe Acute Respiratory Syndrome Coronavirus 2 Through Time. The Journal of Infectious Diseases, 227(1), 9–17. https://doi.org/10.1093/infdis/jiac317
Kanji, J. N., Zelyas, N., MacDonald, C., Pabbaraju, K., Khan, M. N., Prasad, A., Hu, J., Diggle, M., Berenger, B. M., & Tipples, G. (2021). False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virology Journal, 18(1), 13. https://doi.org/10.1186/s12985-021-01489-0
Aggarwal, A., Rani, A., & Kumar, M. (2020). A robust method to authenticate car license plates using segmentation and ROI based approach. Smart and Sustainable Built Environment, 9(4), 737–747. https://doi.org/10.1108/SASBE-07-2019-0083
Aggarwal, G., Jhajharia, K., Izhar, J., Kumar, M., & Abualigah, L. (2023). A Machine Learning Approach to Classify Biomedical Acoustic Features for Baby Cries. Journal of Voice. https://doi.org/10.1016/J.JVOICE.2023.06.014
Alhudhaif, A., Almaslukh, B., Aseeri, A. O., Guler, O., & Polat, K. (2023). A novel nonlinear automated multi-class skin lesion detection system using soft-attention based convolutional neural networks. Chaos, Solitons & Fractals, 170, 113409. https://doi.org/10.1016/j.chaos.2023.113409
Güler, O., & Polat, K. (2022). Classification Performance of Deep Transfer Learning Methods for Pneumonia Detection from Chest X-Ray Images. Journal of Artificial Intelligence and Systems, 4(1), 107–126. https://doi.org/10.33969/AIS.2022040107
Bütüner, R., & Calp, M. H. (2022). Diagnosis and Detection of COVID-19 from Lung Tomography Images Using Deep Learning and Machine Learning Methods. International Journal of Intelligent Systems and Applications in Engineering, 10(2), 190–200. https://ijisae.org/index.php/IJISAE/article/view/1843
Raheja, S., Kasturia, S., Cheng, X., & Kumar, M. (2023). Machine learning-based diffusion model for prediction of coronavirus-19 outbreak. Neural Computing and Applications, 35(19), 13755–13774. https://doi.org/10.1007/s00521-021-06376-x
Al-Saedi, D. K. A., & Savaş, S. (2022). Classification of Skin Cancer with Deep Transfer Learning Method. Computer Science, IDAP-2022(International Artificial Intelligence and Data Processing Symposium), 202–210. https://doi.org/10.53070/BBD.1172782
Madhu, G., Govardhan, A., Ravi, V., Kautish, S., Srinivas, B. S., Chaudhary, T., & Kumar, M. (2022). DSCN-net: a deep Siamese capsule neural network model for automatic diagnosis of malaria parasites detection. Multimedia Tools and Applications, 81(23), 34105–34127. https://doi.org/10.1007/s11042-022-13008-6
Alhatemi, R. A. J., & Savaş, S. (2022). Transfer Learning-Based Classification Comparison of Stroke. Computer Science, IDAP 2022:(International Artificial Intelligence and Data Processing Symposium), 192–201. https://doi.org/10.53070/BBD.1172807
Chen, H., & Sung, J. J. Y. (2021). Potentials of AI in medical image analysis in Gastroenterology and Hepatology. Journal of Gastroenterology and Hepatology, 36(1), 31–38. https://doi.org/10.1111/JGH.15327
Kolla, L., Gruber, F. K., Khalid, O., Hill, C., & Parikh, R. B. (2021). The case for AI-driven cancer clinical trials – The efficacy arm in silico. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 1876(1), 188572. https://doi.org/10.1016/J.BBCAN.2021.188572
Ahsan, M. M., Uddin, M. R., Farjana, M., Sakib, A. N., Momin, K. Al, & Luna, S. A. (2022). Image Data collection and implementation of deep learning-based model in detecting Monkeypox disease using modified VGG16. https://arxiv.org/abs/2206.01862v1
Bala, D., Hossain, M. S., Hossain, M. A., Abdullah, M. I., Rahman, M. M., Manavalan, B., Gu, N., Islam, M. S., & Huang, Z. (2023). MonkeyNet: A robust deep convolutional neural network for monkeypox disease detection and classification. Neural Networks, 161, 757–775. https://doi.org/10.1016/J.NEUNET.2023.02.022
Akın, K. D., Gürkan, Ç., Budak, A., & Karatas, H. (2022). Classification of Monkeypox Skin Lesion using the Explainable Artificial Intelligence Assisted Convolutional Neural Networks. European Journal of Science and Technology, 40, 106–110.
Yaşar, H. (2022). Transfer Derin Öğrenme Kullanılarak Maymun Çiçeği Hastalığının İki Sınıflı ve Çok Sınıflı Sınıflandırılması Üzerine Kapsamlı Bir Çalışma. ELECO 2022 - Elektrik-Elektronik ve Biyomedikal Mühendisliği Konferansı, 1–5.
Haque, Md. E., Ahmed, Md. R., Nila, R. S., & Islam, S. (2022b). Classification of Human Monkeypox Disease Using Deep Learning Models and Attention Mechanisms. ArXiv. https://arxiv.org/abs/2211.15459v1
Dwivedi, M., Tiwari, R. G., & Ujjwal, N. (2022). Deep Learning Methods for Early Detection of Monkeypox Skin Lesion. 2022 8th International Conference on Signal Processing and Communication, ICSC 2022, 343–348. https://doi.org/10.1109/ICSC56524.2022.10009571
Uzun Ozsahin, D., Mustapha, M. T., Uzun, B., Duwa, B., & Ozsahin, I. (2023). Computer-Aided Detection and Classification of Monkeypox and Chickenpox Lesion in Human Subjects Using Deep Learning Framework. Diagnostics, 13(2). https://doi.org/10.3390/diagnostics13020292
Pramanik, R., Banerjee, B., Efimenko, G., Kaplun, D., & Sarkar, R. (2023). Monkeypox detection from skin lesion images using an amalgamation of CNN models aided with Beta function-based normalization scheme. PLOS ONE, 18(4), e0281815. https://doi.org/10.1371/JOURNAL.PONE.0281815
Irmak, M. C., Aydin, T., & Yağanoğlu, M. (2022). Monkeypox Skin Lesion Detection with MobileNetV2 and VGGNet Models. 2022 Medical Technologies Congress (TIPTEKNO), 1–4. https://doi.org/10.1109/TIPTEKNO56568.2022.9960194
Sahin, V. H., Oztel, I., & Yolcu Oztel, G. (2022). Human Monkeypox Classification from Skin Lesion Images with Deep Pre-trained Network using Mobile Application. Journal of Medical Systems, 46(11), 79. https://doi.org/10.1007/s10916-022-01863-7
Sitaula, C., & Shahi, T. B. (2022). Monkeypox Virus Detection Using Pre-trained Deep Learning-based Approaches. Journal of Medical Systems, 46(11), 78. https://doi.org/10.1007/s10916-022-01868-2
Altun, M., Gürüler, H., Özkaraca, O., Khan, F., Khan, J., & Lee, Y. (2023). Monkeypox Detection Using CNN with Transfer Learning. Sensors, 23(4). https://doi.org/10.3390/s23041783
Bala, D., & Hossain, M. S. (2023). Monkeypox Skin Images Dataset (MSID). 6. https://doi.org/10.17632/R9BFPNVYXR.6
Cubuk, E. D., Zoph, B., Shlens, J., & Le, Q. V. (2019). RandAugment: Practical automated data augmentation with a reduced search space. IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, 2020-June, 3008–3017. https://doi.org/10.1109/CVPRW50498.2020.00359
Shorten, C., & Khoshgoftaar, T. M. (2019). A survey on Image Data Augmentation for Deep Learning. Journal of Big Data, 6(1), 1–48. https://doi.org/10.1186/S40537-019-0197-0/FIGURES/33
Pan, S. J., & Yang, Q. (2010). A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering, 22(10), 1345–1359. https://doi.org/10.1109/TKDE.2009.191
Weiss, K., Khoshgoftaar, T. M., & Wang, D. D. (2016). A survey of transfer learning. Journal of Big Data, 3(1), 1–40. https://doi.org/10.1186/S40537-016-0043-6/TABLES/6
Keras. (2023). Keras Applications. Keras Applications. https://keras.io/api/applications/
TensorFlow. (2023). Module: tf.keras.applications | TensorFlow v2.12.0. Module: Tf.Keras.Applications | TensorFlow v2.12.0. https://www.tensorflow.org/api_docs/python/tf/keras/applications
Brown, G. (2010). Ensemble Learning. In G. I. Sammut Claude and Webb (Ed.), Encyclopedia of Machine Learning (pp. 312–320). Springer US. https://doi.org/10.1007/978-0-387-30164-8_252
Deng, L., & Yu, D. (2013). Deep learning: Methods and applications. Foundations and Trends in Signal Processing, 7(3–4), 197–387. https://doi.org/10.1561/2000000039
Polikar, R. (2006). Ensemble based systems in decision making. IEEE Circuits and Systems Magazine, 6(3), 21–44. https://doi.org/10.1109/MCAS.2006.1688199
Ng, K. W., Tian, G. L., & Tang, M. L. (2011). Dirichlet and Related Distributions: Theory, Methods and Applications. In Dirichlet and Related Distributions: Theory, Methods and Applications. John Wiley & Sons, Ltd. https://doi.org/10.1002/9781119995784
Borges, J. (2019). DeepStack: Ensembles for Deep Learning. https://github.com/jcborges/DeepStack
Howard, J., & Ruder, S. (2018). Universal Language Model Fine-tuning for Text Classification. ACL 2018 - 56th Annual Meeting of the Association for Computational Linguistics, Proceedings of the Conference (Long Papers), 1, 328–339. https://doi.org/10.18653/v1/p18-1031
Yosinski, J., Clune, J., Bengio, Y., & Lipson, H. (2014). How transferable are features in deep neural networks? Advances in Neural Information Processing Systems, 4(January), 3320–3328. https://arxiv.org/abs/1411.1792v1
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., & Salakhutdinov, R. (2014). Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Journal of Machine Learning Research, 15(56), 1929–1958. http://jmlr.org/papers/v15/srivastava14a.html
Gao, B., & Pavel, L. (2017). On the Properties of the Softmax Function with Application in Game Theory and Reinforcement Learning. https://arxiv.org/abs/1704.00805v4
Bock, S., Goppold, J., & Weiß, M. (2018). An improvement of the convergence proof of the ADAM-Optimizer. https://arxiv.org/abs/1804.10587v1
Kingma, D. P., & Ba, J. L. (2014). Adam: A Method for Stochastic Optimization. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. https://arxiv.org/abs/1412.6980v9
Goodfellow, I., Bengio, Y., & Courville, A. (2016). Deep learning. MIT press.
Gómez, R. (2018). Understanding Categorical Cross-Entropy Loss, Binary Cross-Entropy Loss, Softmax Loss, Logistic Loss, Focal Loss and all those confusing names. Github. https://gombru.github.io/2018/05/23/cross_entropy_loss/
WHO. (2022). Second meeting of the International Health Regulations (2005) (IHR) Emergency Committee regarding the multi-country outbreak of monkeypox. Second Meeting of the International Health Regulations (2005) (IHR) Emergency Committee Regarding the Multi-Country Outbreak of Monkeypox. https://www.who.int/news/item/23-07-2022-second-meeting-of-the-international-health-regulations-(2005)-(ihr)-emergency-committee-regarding-the-multi-country-outbreak-of-monkeypox
Nuzzo, J. B., Borio, L. L., & Gostin, L. O. (2022). The WHO Declaration of Monkeypox as a Global Public Health Emergency. JAMA, 328(7), 615–617. https://doi.org/10.1001/JAMA.2022.12513
Author information
Authors and Affiliations
Contributions
Serkan Savaş: conceptualization, methodology, investigation, data acquisition, testing, validation, visualization, writing, original draft, and editing.
Corresponding author
Ethics declarations
Ethics Approval
Only data coming from publicly available datasets were used and ethics approval not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Savaş, S. Enhancing Disease Classification with Deep Learning: a Two-Stage Optimization Approach for Monkeypox and Similar Skin Lesion Diseases. J Digit Imaging. Inform. med. 37, 778–800 (2024). https://doi.org/10.1007/s10278-023-00941-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00941-7