Abstract
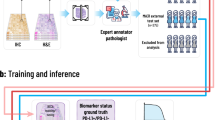
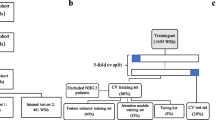
In prognostic evaluation of breast cancer, immunohistochemical (IHC) marker human epidermal growth factor receptor 2 (HER2) is used for prognostic evaluation. Accurate assessment of HER2-stained tissue sample is essential in therapeutic decision making for the patients. In regular clinical settings, expert pathologists assess the HER2-stained tissue slide under microscope for manual scoring based on prior experience. Manual scoring is time consuming, tedious, and often prone to inter-observer variation among group of pathologists. With the recent advancement in the area of computer vision and deep learning, medical image analysis has got significant attention. A number of deep learning architectures have been proposed for classification of different image groups. These networks are also used for transfer learning to classify other image classes. In the presented study, a number of transfer learning architectures are used for HER2 scoring. Five pre-trained architectures viz. VGG16, VGG19, ResNet50, MobileNetV2, and NASNetMobile with decimating the fully connected layers to get 3-class classification have been used for the comparative assessment of the networks as well as further scoring of stained tissue sample image based on statistical voting using mode operator. HER2 Challenge dataset from Warwick University is used in this study. A total of 2130 image patches were extracted to generate the training dataset from 300 training images corresponding to 30 training cases. The output model is then tested on 800 new test image patches from 100 test images acquired from 10 test cases (different from training cases) to report the outcome results. The transfer learning models have shown significant accuracy with VGG19 showing the best accuracy for the test images. The accuracy is found to be 93%, which increases to 98% on the image-based scoring using statistical voting mechanism. The output shows a capable quantification pipeline in automated HER2 score generation.




Similar content being viewed by others
References
Ma J, Jemal A: Breast cancer statistics: Springer, 2013
Mosquera-Lopez C, Agaian S, Velez-Hoyos A, Thompson I, et al: Computer-aided prostate cancer diagnosis from digitized histopathology: a review on texture-based systems. IEEE reviews in biomedical engineering 8:98-113, 2014
Joensuu K, Leidenius M, Kero M, Andersson LC, Horwitz KB, Heikkilä P, et al: ER, PR, HER2, Ki-67 and CK5 in early and late relapsing breast cancer--reduced CK5 expression in metastases. Breast cancer: basic and clinical research 7:23, 2013
Wolff AC, et al: Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Archives of Pathology and Laboratory Medicine 138:241-256, 2014
Rakha EA, et al: Updated UK Recommendations for HER2 assessment in breast cancer. Journal of clinical pathology 68:93-99, 2015
Nitta H, et al: The assessment of HER2 status in breast cancer: the past, the present, and the future. Pathology international 66:313-324, 2016
Qaiser T, et al: Her 2 challenge contest: a detailed assessment of automated her 2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 72:227-238, 2018
Cordeiro CQ, Ioshii SO, Alves JH, Oliveira LF et al: An Automatic Patch-based Approach for HER-2 Scoring in Immunohistochemical Breast Cancer Images Using Color Features. arXiv preprint https://arxiv.org/1805.05392, 2018
Jeung J, Patel R, Vila L, Wakefield D, Liu C et al: Quantitation of HER2/neu expression in primary gastroesophageal adenocarcinomas using conventional light microscopy and quantitative image analysis. Archives of pathology & laboratory medicine 136:610-617, 2012
Brügmann A, et al: Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains. Breast cancer research and treatment 132:41-49, 2012
Dobson L, et al: Image analysis as an adjunct to manual HER‐2 immunohistochemical review: a diagnostic tool to standardize interpretation. Histopathology 57:27-38, 2010
Ruifrok AC, Johnston DA: Quantification of histochemical staining by color deconvolution. Analytical and quantitative cytology and histology 23:291-299, 2001
Tuominen VJ, Tolonen TT, Isola J et al: ImmunoMembrane: a publicly available web application for digital image analysis of HER2 immunohistochemistry. Histopathology 60:758-767, 2012
Tabakov M, Kozak P: Segmentation of histopathology HER2/neu images with fuzzy decision tree and Takagi–Sugeno reasoning. Computers in biology and medicine 49:19-29, 2014
Pham N-A, et al: Quantitative image analysis of immunohistochemical stains using a CMYK color model. Diagnostic pathology 2:1, 2007
Wdowiak M, Markiewicz T, Osowski S, Swiderska Z, Patera J, Kozlowski W, et al: Hourglass shapes in rank grey-level hit-or-miss transform for membrane segmentation in HER2/neu images. Proc. International Symposium on Mathematical Morphology and Its Applications to Signal and Image Processing: City
Gavrielides MA, Masmoudi H, Petrick N, Myers KJ, Hewitt SM, et al: Automated evaluation of HER-2/neu immunohistochemical expression in breast cancer using digital microscopy. Proc. 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro: City
Wan T, Cao J, Chen J, Qin Z, et al: Automated grading of breast cancer histopathology using cascaded ensemble with combination of multi-level image features. Neurocomputing 229:34-44, 2017
Mukundan R: Analysis of Image Feature Characteristics for Automated Scoring of HER2 in Histology Slides. Journal of Imaging 5:35, 2019
Tewary S, Arun I, Ahmed R, Chatterjee S, Mukhopadhyay S, et al: AutoIHC‐Analyzer: computer‐assisted microscopy for automated membrane extraction/scoring in HER2 molecular markers. Journal of Microscopy 281:87-96, 2021
LeCun Y, Bengio Y, Hinton G, et al: Deep learning. nature 521:436-444, 2015
Huang Y, Zheng H, Liu C, Ding X, Rohde GK, et al: Epithelium-stroma classification via convolutional neural networks and unsupervised domain adaptation in histopathological images. IEEE journal of biomedical and health informatics 21:1625-1632, 2017
Meng N, Lam EY, Tsia KK, So HK-H, et al: Large-scale multi-class image-based cell classification with deep learning. IEEE journal of biomedical and health informatics 23:2091-2098, 2018
Pitkäaho T, Lehtimäki TM, McDonald J, Naughton TJ, et al: Classifying HER2 breast cancer cell samples using deep learning. Proc. Proc Irish Mach Vis Image Process Conf: City
Vandenberghe ME, Scott ML, Scorer PW, Söderberg M, Balcerzak D, Barker C, et al: Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer. Scientific reports 7:45938, 2017
Saha M, Chakraborty C: Her2net: A deep framework for semantic segmentation and classification of cell membranes and nuclei in breast cancer evaluation. IEEE Transactions on Image Processing 27:2189-2200, 2018
Khameneh FD, Razavi S, Kamasak M, et al: Automated segmentation of cell membranes to evaluate HER2 status in whole slide images using a modified deep learning network. Computers in biology and medicine, 2019
Qaiser T, Rajpoot NM: Learning where to see: A novel attention model for automated immunohistochemical scoring. IEEE transactions on medical imaging 38:2620-2631, 2019
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L, et al: Imagenet: A large-scale hierarchical image database. Proc. IEEE conference on computer vision and pattern recognition: City,2009
Huh M, Agrawal P, Efros AA, et al: What makes ImageNet good for transfer learning? arXiv preprint https://arxiv.org/1608.08614, 2016
Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I, et al: Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 27:317-328, 2018
Cheng PM, Malhi HS: Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. Journal of digital imaging 30:234-243, 2017
He K, Zhang X, Ren S, Sun J, et al: Deep residual learning for image recognition. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C, et al: Mobilenetv2: Inverted residuals and linear bottlenecks. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Zoph B, Vasudevan V, Shlens J, Le QV, et al: Learning transferable architectures for scalable image recognition. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint https://arxiv.org/1409.1556, 2014
Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J, et al: Mitosis detection in breast cancer histology images with deep neural networks. Proc. International conference on medical image computing and computer-assisted intervention: City
Khan S, Islam N, Jan Z, Din IU, Rodrigues JJC, et al: A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognition Letters 125:1-6, 2019
Sokolova M, Lapalme G: A systematic analysis of performance measures for classification tasks. Information processing & management 45:427-437, 2009
Acknowledgements
The authors would like to thank the HER2 Challenge research group in the Department of Computer Science, University of Warwick, UK for the access to Challenge dataset. Suman Tewary is grateful to Director, CSIR-Central Scientific Instruments Organisation, Chandigarh for providing the research facility.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tewary, S., Mukhopadhyay, S. HER2 Molecular Marker Scoring Using Transfer Learning and Decision Level Fusion. J Digit Imaging 34, 667–677 (2021). https://doi.org/10.1007/s10278-021-00442-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-021-00442-5