Abstract
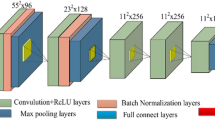
Although magnetic resonance imaging (MRI) has a higher sensitivity of early breast cancer than mammography, the specificity is lower. The purpose of this study was to develop a computer-aided diagnosis (CAD) scheme for distinguishing between benign and malignant breast masses on dynamic contrast material-enhanced MRI (DCE-MRI) by using a deep convolutional neural network (DCNN) with Bayesian optimization. Our database consisted of 56 DCE-MRI examinations for 56 patients, each of which contained five sequential phase images. It included 26 benign and 30 malignant masses. In this study, we first determined a baseline DCNN model from well-known DCNN models in terms of classification performance. The optimum architecture of the DCNN model was determined by changing the hyperparameters of the baseline DCNN model such as the number of layers, the filter size, and the number of filters using Bayesian optimization. As the input of the proposed DCNN model, rectangular regions of interest which include an entire mass were selected from each of DCE-MRI images by an experienced radiologist. Three-fold cross validation method was used for training and testing of the proposed DCNN model. The classification accuracy, the sensitivity, the specificity, the positive predictive value, and the negative predictive value were 92.9% (52/56), 93.3% (28/30), 92.3% (24/26), 93.3% (28/30), and 92.3% (24/26), respectively. These results were substantially greater than those with the conventional method based on handcrafted features and a classifier. The proposed DCNN model achieved high classification performance and would be useful in differential diagnoses of masses in breast DCE-MRI images as a diagnostic aid.




Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A, et al: Cancer statistics, 2017. CA: a cancer journal for clinicians 2017; 67(1): 7–30
Reynolds HE, Jackson VP: Self-referred mammography patients: analysis of patients' characteristics. American Journal of Roentgenology 1991; 157: 481-484
Kuhl CK, Schild HH: Dynamic image interpretation of MRI of the breast. Journal of Magnetic Resonance Imaging 2000; 12(6): 965-974
Gubern-Mérida A, Martí R, Melendez J, Hauth JL, Mann RM, Karssemeijer N, Platel B, et al: Automated localization of breast cancer in DCE-MRI. Medical image analysis 2015; 20(1): 265-274
Leach MO, Boggis CR, Dixon AK, et al: Screening with magnetic resonance imaging and mammography of a UK population at high familial risk of breast cancer: a prospective multicentre cohort study (MARIBS), LANCET 2005; 365: 1769-1778
Peters NH, Borel Rinkes IH, Zuithoff NP, Mali WP, Moons KG, Peeters PH, et al: Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology 2008; 246(1): 116-124
Sardanelli F, Podo F, D'Agnolo G, et al: Multicenter comparative multimodality surveillance of women at genetic-familial high risk for breast cancer (HIBCRIT study): interim results. Radiology 2007; 242(3): 698-715
Kuhl CK, Schrading S, Leutner CC, et al: Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. Journal of Clinical Oncology 2005; 23: 8469–8476
Pisano ED, Hendrick RE, Yaffe MJ, Baum JK, Acharyya S, Cormack JB, D'Orsi CJ, et al: Diagnostic accuracy of digital versus film mammography: exploratory analysis of selected population subgroups in DMIST. Radiology 2008; 246(2): 376-383
Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY: Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Academic Radiology 2008; 15(12): 1513–1525
Siegmann KC, Kra¨mer B, Claussen C, et al: Current status and new developments in breast MRI. Breast Care (Basel) 2011; 6(2): 87–92
Fusco R, Sansone M, Filice S, Carone G, Amato DM, Sansone C, Petrillo A, et al: Pattern recognition approaches for breast cancer DCE-MRI classification: a systematic review. Journal of medical and biological engineering 2016; 36(4): 449-459
Berg WA, Blume JD, Cormack JB, et al.: Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer. The Journal of the American Medical Association 2008; 299(18): 2151-2163
Chae EY, Kim HH, Cha JH, Shin HJ, Kim H, et al: Evaluation of Screening Whole‐Breast Sonography as a Supplemental Tool in Conjunction with Mammography in Women With Dense Breasts. Journal of Ultrasound in Medicine 2013; 32(9): 1573-1578
Doi K, MacMahon H, Katsuragawa S, Nishikawa RM, Jiang Y, et al: Computer-aided diagnosis in radiology: potential and pitfalls. European journal of Radiology 1999; 31(2): 97–109
Doi K: Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Computerized medical imaging and graphics 2007; 31(4–5): 198–211
Hizukuri A, Nakayama R, Kashikura Y, Takase H, Kawanaka H, Ogawa T, Tsuruoka S, et al: Computerized determination scheme for histological classification of breast mass using objective features corresponding to clinicians’ subjective impressions on ultrasonographic images. Journal of digital imaging 2013; 26(5): 958-970
Hizukuri A, Nakayama R: Computer-Aided Diagnosis Scheme for Determining Histological Classification of Breast Lesions on Ultrasonographic Images Using Convolutional Neural Network. Diagnostics 2018; 8(3): 1-9
Honda E, Nakayama R, Koyama H, Yamashita A, et al: Computer-aided diagnosis scheme for distinguishing between benign and malignant masses in breast DCE-MRI. Journal of digital imaging 2016; 29(3): 388-393
Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 2012; 1097–1105
Zeiler MD, Fergus R: Visualizing and understanding convolutional networks. In European conference on computer vision, Springer, Cham, 2014; 818-833
Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. Int. Conf. on Learning Representations 2015; San Diego, CA
Christian S, et.al.: Going deeper with convolutions, Proceedings of the IEEE conference on computer vision and pattern recognition. 2015
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L, et al: ImageNet Large Scale Visual Recognition Challenge, International Journal of Computer Vision, 2015; 115(3); 211-252
LeCun Y, Bengio Y, Hinton G, et al: Deep learning. nature 2015; 521(7553): 436
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Sánchez CI, et al: A survey on deep learning in medical image analysis, Medical image analysis 2017; 42: 60-88
Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM, Giger ML, et al: Deep learning in medical imaging and radiation therapy, Medical Physics 2019; 46(1)
Bayramoglu N, Juho K, Janne H, et al: Deep learning for magnification independent breast cancer histopathology image classification, 23rd International conference on pattern recognition (ICPR), 2016; 2440–2445
Jadoon MM, Zhang Q, Haq IU, Butt S, Jadoon A, et al: Three-class mammogram classification based on descriptive CNN features, BioMed Research International, 2017, 1–11
Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M, et al: Medical image classification with convolutional neural network, 13th International Conference on Control Automation Robotics & Vision (ICARCV), 2014, 844–848
Nejad EM, Affendey LS, Latip RB, Bin Ishak, et al: Classification of histopathology images of breast into benign and malignant using a single-layer convolutional neural network, Proceedings of the International Conference on Imaging, Signal Processing and Communication, 2017, 50–53
Rasmussen CE: Gaussian processes in machine learning. In Summer School on Machine Learning, Springer, Berlin, Heidelberg, 2003, 63-71
Le HT, Phung SL, Bouzerdoum A, Tivive FHC, et al: Human motion classification with micro-doppler radar and bayesian-optimized convolutional neural networks. In 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) , 2018, 2961–2965
Snoek J, Larochelle H, Adams RP: Practical Bayesian optimization of machine learning algorithms. In Advances in Neural Information Processing Systems, 2012, 2951–2959
Dernoncourt F, Lee JY: Optimizing neural network hyperparameters with Gaussian processes for dialog act classification. In 2016 IEEE Spoken Language Technology Workshop (SLT), 2016, 406–413
Metz CE: ROC methodology in radiologic imaging. Investigative Radiology 1986; 21: 720–733
Peng Y, Jiang Y, Antic T, Giger ML, Eggener SE, Oto A, et al: Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology 2014; 271(2): 461-471
Cireşan DC, Meier U, Gambardella LM, Schmidhuber J, et al: Deep, big, simple neuralnets for handwritten digit recognition. Neural computation 2010; 22(12): 3207-3220
Langlotz CP: Fundamental measures of diagnostic examination performance: Usefulness for clinicaldecision making and research. Radiology 2003; 228: 3–9
Otsu N: A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics 1979; 9(1): 62–66
Nakayama R, Uchiyama Y, Watanabe R, Katsuragawa S, Namba K, Doi K, et al: Computer‐aided diagnosis scheme for histological classification of clustered microcalcifications on magnification mammograms. Medical physics 2004; 31(4): 789–799
Nakayama R, Watanabe R, Namba K, Takeda K, Yamamoto K, Katsuragawa S, Doi K, et al: Computer-aided diagnosis scheme for identifying histological classification of clustered microcalcifications by use of follow-up magnification mammograms. Academic radiology 2006; 13(10): 1219–1228
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hizukuri, A., Nakayama, R., Nara, M. et al. Computer-Aided Diagnosis Scheme for Distinguishing Between Benign and Malignant Masses on Breast DCE-MRI Images Using Deep Convolutional Neural Network with Bayesian Optimization. J Digit Imaging 34, 116–123 (2021). https://doi.org/10.1007/s10278-020-00394-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00394-2