Abstract
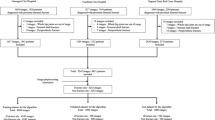
To use deep learning with advanced data augmentation to accurately diagnose and classify femoral neck fractures. A retrospective study of patients with femoral neck fractures was performed. One thousand sixty-three AP hip radiographs were obtained from 550 patients. Ground truth labels of Garden fracture classification were applied as follows: (1) 127 Garden I and II fracture radiographs, (2) 610 Garden III and IV fracture radiographs, and (3) 326 normal hip radiographs. After localization by an initial network, a second CNN classified the images as Garden I/II fracture, Garden III/IV fracture, or no fracture. Advanced data augmentation techniques expanded the training set: (1) generative adversarial network (GAN); (2) digitally reconstructed radiographs (DRRs) from preoperative hip CT scans. In all, 9063 images, real and generated, were available for training and testing. A deep neural network was designed and tuned based on a 20% validation group. A holdout test dataset consisted of 105 real images, 35 in each class. Two class prediction of fracture versus no fracture (AUC 0.92): accuracy 92.3%, sensitivity 0.91, specificity 0.93, PPV 0.96, NPV 0.86. Three class prediction of Garden I/II, Garden III/IV, or normal (AUC 0.96): accuracy 86.0%, sensitivity 0.79, specificity 0.90, PPV 0.80, NPV 0.90. Without any advanced augmentation, the AUC for two-class prediction was 0.80. With DRR as the only advanced augmentation, AUC was 0.91 and with GAN only AUC was 0.87. GANs and DRRs can be used to improve the accuracy of a tool to diagnose and classify femoral neck fractures.




Similar content being viewed by others
References
Papadimitriou N, Tsilidis KK, Orfanos P, Benetou V, Ntzani EE, Soerjomataram I, et al. Burden of hip fracture using disability-adjusted life-years: a pooled analysis of prospective cohorts in the CHANCES consortium. Lancet Public Heal [Internet]. 2017 [cited 2018 Sep 13];2(5):e239–46. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29253489
Kani KK, Porrino JA, Mulcahy H, Chew FS. Fragility fractures of the proximal femur: review and update for radiologists. Skeletal Radiol [Internet]. 2018 Jun 29 [cited 2018 Sep 13]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/29959502
Sozen T, Ozisik L, Calik Basaran N. An overview and management of osteoporosis. Eur J Rheumatol [Internet]. 2017 [cited 2018 Sep 13];4(1):46–56. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28293453
Natasha Morrissey, Efthymios Iliopoulos, Ahmad Wais Osmani, Kevin Newman. Injury, Int. J. Care Injured. 2017; 48: 1155–1158.
Ryan DJ, Yoshihara H, Yoneoka D, Egol KA, Zuckerman JD. Delay in hip fracture surgery. J Orthop Trauma [Internet]. 2015 [cited 2018 Sep 13];29(8):343–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25714442
Garden RS. Low-angle fixation in fractures of the femoral neck. J Bone Joint Surg Br [Internet]. The British Editorial Society of Bone and Joint Surgery; 1961 1 [cited 2018 Sep 13];43–B(4):647–63. Available from: https://doi.org/10.1302/0301-620X.43B4.647
Florschutz A V., Langford JR, Haidukewych GJ, Koval KJ. Femoral Neck fractures. J Orthop Trauma [Internet]. 2015 [cited 2018 Sep 13];29(3):121–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25635363
Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE [Internet]. 1998 [cited 2018 13];86(11):2278–324. Available from: http://ieeexplore.ieee.org/document/726791/
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature [Internet]. Nature Publishing Group; 2017 25 [cited 2018 Sep 13];542(7639):115–8. Available from: http://www.nature.com/articles/nature21056
Chi J, Walia E, Babyn P, Wang J, Groot G, Eramian M. Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network. J Digit Imaging [Internet] 2017 Aug 10 [cited 2018 Sep 13];30(4):477–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28695342
Gale W, Oakden-Rayner L, Carneiro G, Bradley AP, Palmer LJ. Detecting hip fractures with radiologist-level performance using deep neural networks. 2017 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1711.06504
Kazi A, Albarqouni S, Sanchez AJ, Kirchhoff S, Biberthaler P, Navab N, et al. Automatic classification of proximal femur fractures based on attention models. In Springer, Cham; 2017 [cited 2018 Sep 13]. p. 70–8. Available from: https://doi.org/10.1007/978-3-319-67389-9_9
Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks [Internet]. [cited 2018 Sep 13]. Available from: http://www.iro.umontreal.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2016 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1610.02391
Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. Striving for simplicity: the all convolutional net. 2014 21 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1412.6806
Tian TP, Chen Y, Leow WK, Hsu W, Howe T Sen, Png MA. Computing neck-shaft angle of femur for x-ray fracture detection. In Springer, Berlin, Heidelberg; 2003 [cited 2018 Sep 13]. p. 82–9. Available from: https://doi.org/10.1007/978-3-540-45179-2_11
Zhou J, Chan KL, Chong VFH, Krishnan SM. Extraction of brain tumor from MR images using one-class support vector machine. In: 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference [Internet]. IEEE; 2005 [cited 2018 Sep 13]. p. 6411–4. Available from: http://ieeexplore.ieee.org/document/1615965/
Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging [Internet]. 2011 [cited 2018 Sep 13];33(2):296–305. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21274970
Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Confounding variables can degrade generalization performance of radiological deep learning models. 2018 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1807.00431
Urakawa T, Tanaka Y, Goto S, Matsuzawa H, Watanabe K, Endo N. Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skeletal Radiol [Internet]. 2019 [cited 2019 Jan 16];48(2):239–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29955910
Dominguez S, Liu P, Roberts C, Mandell M, Richman PB. Prevalence of traumatic hip and pelvic fractures in patients with suspected hip fracture and negative initial standard radiographs—a study of emergency department patients. Academic emergency medicine. 2005 Apr;12(4):366-9.
Appendix References
Arjovsky M, Chintala S, Bottou L. Wasserstein GAN 2017 Jan 26 [cited 2018]; Available from: http://arxiv.org/abs/1701.07875
Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, Courville A. Improved Training of Wasserstein GANs Montreal Institute for Learning Algorithms [Internet]. [cited 2018 Sep 13]. Available from: https://github.com/igul222/improved_wgan_training.
He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2015 10 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1512.03385
Jaderberg M, Simonyan K, Zisserman A, Kavukcuoglu K. Spatial Transformer Networks. 2015 5 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1506.02025
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going Deeper with Convolutions. 2014 16 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1409.4842
Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. 2015 [cited 2018 Sep 13]; Available from: http://arxiv.org/abs/1502.03167
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J Mach Learn Res [Internet]. 2014 [cited 2018 13];15:1929–58. Available from: http://jmlr.org/papers/v15/srivastava14a.html
Dozat T. Incorporating Nesterov Momentum into Adam [Internet]. [cited 2018 Sep 13]. Available from: http://mattmahoney.net/dc/text8.zip
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Data Augmentation
Data augmentation used in this study entailed several real-time modifications to the source images at the time of training. These modifications included (1) random horizontal and vertical flipping of the input image; (2) random rotation of the input image by − 30° to 30°; and (3) random contrast jittering of the input image and addition of a random Gaussian noise matrix, performed to simulate different acquisition parameters for each image. This resulted in roughly an additional 3.2 × 108 variations of each input image.
For further data augmentation, 6000 digitally reconstructed radiographs (DRRs) were generated. DRR volume rendering, also called simulated x-ray volume rendering, is a direct volume rendering technique that consists of simulating x-rays passing through the reconstructed CT volume based on the absorption properties of the tissue. DRR generation is a popular technique in simulating radiation therapy treatments. For this project, we recreated a cone-beam radiographic acquisition with a ray-tracing DRR generating algorithm utilizing the thin slice acquisitions, where available. Forty-five hip CT volumes from 34 patients (11 patients had a pelvic CT scan performed which contributed two hips) were obtained that were performed within 3 days of the hip radiographs but before surgery. Using 3D input volumes allowed for a far greater range of augmentation capabilities. We applied rigid affine transformation and multiple slightly different acquisition parameters, allowing us to effectively turn one input example into multiple slightly different radiographs. We were able to take one patient volume and simulate (1) making the patient slightly thinner, larger, taller or shorter; (2) varying radiographic KvP, mA, and source to object distance; and (3) simulating internal/external rotation of the hip and minor variations in apical angulation of the patient.
Additional training examples were generated utilizing a generative adversarial network (GAN). GANs are a family of deep generative models which balance training of a generative network and a discriminative network in order to generate realistic examples based on a training set data distributions. To summarize our design choices in this paper, a deep convolutional GAN with a residual network generator was utilized. Earthmover distance based on the paper by Arjovsky et al. [22] was used as the discriminatory function with gradient clipping. The GAN was trained for 300 epochs which took 72 h on the research computer until convergence. Then, 2000 generated outputs were subsequently used as an additional input into the network. All artificially generated images (using DRR and GAN techniques) were utilized as additional data for training only, and not for testing or validation.
Neural Network Implementation Details
The overall network architecture is shown in Table 1 and Fig. 2. The CNN was implemented by a series of 3 × 3 convolutional kernels to maximize computational efficiency while preserving nonlinearity [23]. After an initial standard convolutional layer, a series of residual layers are utilized in the network. Originally described by He et al. [24], residual neural networks can stabilize gradients during backpropagation, leading to improved optimization and facilitating greater network depth. A spatial transformer module was inserted after the 11th hidden layer. Initially applied to convolutional neural networks by Jaderberg et al. [25], spatial transformer layers allow a network to explicitly learn affine transformation parameters that regularize global spatial variations in feature space. This is important for the network to be robust against significant variations in imaging technique or positioning commonly seen in practice.
Downsampling of feature map size was implemented by means of strided convolutions. All nonlinear functions utilize the rectified linear unit (ReLU) which allows training of deep neural networks by stabilizing gradients on backpropagation [26]. Additionally, batch normalization was used between the convolutional and ReLU layers to prevent covariate shift [27]. Upon downsampling, the number of feature channels is doubled, preventing a representation bottleneck. Dropout with a keep probability of 75% was applied to the first fully connected layer to limit over-fitting and add stochasticity to the training process [28].
In addition to the customized network described above, several additional network architectures were tested. This includes (1) ResNet 52 network architecture initialized both randomly and with pre-trained weights from Imagenet; (2) custom-built networks, initialized from random weights, and with varying numbers of convolutional layers based on the Inception v4 architecture; and (3) 100 layer network based on a randomly initialized DenseNet architecture. Performance for the networks was best when initializing weights randomly across the board. We found that when using greater than 14 hidden layers in two dimensional residual networks overfitting occurred.
Training was implemented using the parameterized Adam optimizer, combined with the Nesterov accelerated gradient described by Dozat [29]. Parameters were initialized to equalize input and output variance utilizing the heuristic described by Glorot et al. [13]. L2 regularization was implemented to prevent overfitting of data by limiting the squared magnitude of the kernel weights. Final hyperparameter settings included a learning rate set to 1e-3, keep probability for dropout of 50%, moving average weight decay of 0.999, and L2 regularization weighting of 1e-4.
Software code for this study was written in Python (v3.5) using the TensorFlow module (v1.5). Experiments and CNN training were performed on a Linux workstation with NVIDIA Titan X Pascal GPU with 12 GB on chip memory, i7 CPU and 32 GB RAM. The classification network was trained for 200 epochs, taking 5 h. Inference time was 22 s per image for localization and classification. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
Rights and permissions
About this article
Cite this article
Mutasa, S., Varada, S., Goel, A. et al. Advanced Deep Learning Techniques Applied to Automated Femoral Neck Fracture Detection and Classification. J Digit Imaging 33, 1209–1217 (2020). https://doi.org/10.1007/s10278-020-00364-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00364-8