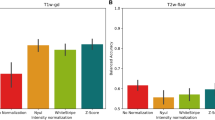
Abstract
The growing interest in machine learning (ML) in healthcare is driven by the promise of improved patient care. However, how many ML algorithms are currently being used in clinical practice? While the technology is present, as demonstrated in a variety of commercial products, clinical integration is hampered by a lack of infrastructure, processes, and tools. In particular, automating the selection of relevant series for a particular algorithm remains challenging. In this work, we propose a methodology to automate the identification of brain MRI sequences so that we can automatically route the relevant inputs for further image-related algorithms. The method relies on metadata required by the Digital Imaging and Communications in Medicine (DICOM) standard, resulting in generalizability and high efficiency (less than 0.4 ms/series). To support our claims, we test our approach on two large brain MRI datasets (40,000 studies in total) from two different institutions on two different continents. We demonstrate high levels of accuracy (ranging from 97.4 to 99.96%) and generalizability across the institutions. Given the complexity and variability of brain MRI protocols, we are confident that similar techniques could be applied to other forms of radiological imaging.











Similar content being viewed by others
References
Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, Geis JR, Pandharipande PV, Brink JA, Dreyer KJ: Current applications and future impact of machine learning in radiology. Radiology 288(2):318–328, 2018
Koohy H: The Rise and Fall of Machine Learning Methods in Biomedical Research. F1000Research 6:2012, 2018
Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, Wang Y, Dong Q, Shen H, Wang Y: Artificial intelligence in healthcare: past, present and future. Stroke and Vascular Neurology 2(4):230–243, 2017
Allen B et al.: A road map for translational research on artificial intelligence in medical imaging: from the 2018 National Institutes of Health/RSNA/ACR/the Academy Workshop. J Am Coll Radiol 16(9):1179–1189, 2019
DICOM standard. [Online]. Available: https://www.dicomstandard.org/. [Accessed: 20-Sep-2018].
Petrakis EGM, Faloutsos A: Similarity searching in medical image databases. IEEE Trans Knowl Data Eng 9(3):435–447, 1997
Lehmann TM, Schubert H, Keysers D, Kohnen M, Wein BB: The IRMA Code for Unique Classification of Medical Images, presented at the Medical Imaging. San Diego 2003, p 440
M. O. Gueld et al., Quality of DICOM Header Information for Image Categorization, presented at the Medical Imaging 2002, San Diego 280–287.
Bergamasco LCC, Nunes FLS: Intelligent retrieval and classification in three-dimensional biomedical images — a systematic mapping. Comput Sci Rev 31:19–38, 2019
Kwak D-M, Kim B-S, Yoon O-K, Park C-H, Won J-U, Park K-H: Content-based ultrasound image retrieval using a coarse to fine approach. Ann NY Acad Sci 980(1):212–224, 2002
Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H: Visualizing and Enhancing a Deep Learning Framework Using Patients Age and Gender for Chest X-ray Image Retrieval, presented at the SPIE Medical Imaging, San Diego 2016, p 978510
Stanley RJ, De S, Demner-Fushman D, Antani S, Thoma GR: An image feature-based approach to automatically find images for application to clinical decision support. Computerized Medical Imaging and Graphics 35(5):365–372, 2011
Quellec G, Lamard M, Cazuguel G, Roux C, Cochener B: Case retrieval in medical databases by fusing heterogeneous information. IEEE Trans Med Imaging 30(1):108–118, 2011
de Herrera AGS, Schaer R, Bromuri S, Muller H: Overview of the ImageCLEF 2016 medical task, in Working Notes of CLEF 2016 (Cross Language Evaluation Forum), 2016.
de Herrera AGS, Markonis D, Müller H: Bag-of-colors for biomedical document image classification. In: Greenspan H, Müller H, Syeda-Mahmood T Eds. Medical Content-Based Retrieval for Clinical Decision Support, Vol. 7723. Berlin: Springer Berlin Heidelberg, 2013, pp. 110–121
Cirujeda P, Binefa X: Medical Image Classification via 2D Color Feature Based Covariance Descriptors, Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015, 2015, p. 10
Pelka O, Friedrich CM: FHDO Biomedical Computer Science Group at Medical Classification Task of Image CLEF 2015, Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015, 2015, p. 15
Kumar A, Kim J, Lyndon D, Fulham M, Feng D: An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inf 21(1):31–40, 2017
Koitka S, Friedrich CM: Traditional Feature Engineering and Deep Learning Approaches at Medical Classification Task of Image CLEF 2016. CLEF, 2016, p. 15
Quddus A, Basir O: Semantic image retrieval in magnetic resonance brain volumes. IEEE Transactions on Information Technology in Biomedicine 16(3):348–355, 2012
Müller H, Michoux N, Bandon D, Geissbuhler A: A review of content-based image retrieval systems in medical applications—clinical benefits and future directions. International Journal of Medical Informatics 73(1):1–23, Feb. 2004
Mohanapriya S, Vadivel M: Automatic retrieval of MRI brain image using multiqueries system, in 2013 International Conference on Information Communication and Embedded Systems (ICICES), Chennai, 2013, pp 1099–1103.
Li Z, Zhang X, Müller H, Zhang S: Large-scale retrieval for medical image analytics: a comprehensive review. Medical Image Analysis 43:66–84, 2018
Müller H, Rosset A, Vallée J-P, Geissbuhler A: Integrating content-based visual access methods into a medical case database. Studies in Health Technology and Informatics 95:6, 2003
Caicedo JC, Gonzalez FA, Romero E: A semantic content-based retrieval method for histopathology images. In: Li H, Liu T, Ma W-Y, Sakai T, Wong K-F, Zhou G Eds. Information Retrieval Technology, Vol. 4993. Berlin: Springer Berlin Heidelberg, 2008, pp. 51–60
C. Brodley, A. Kak, C. Shyu, J. Dy, L. Broderick, and A. M. Aisen, Content-Based Retrieval from Medical Image Databases: a Synergy of Human Interaction, Machine Learning and Computer Vision. In: AAAI ‘99 Proceedings of the Sixteenth National Conference on Artificial Intelligence and the Eleventh Innovative Applications of Artificial Intelligence Conference Innovative Applications of Artificial Intelligence, 1999, pp 760–767.
Mattie ME, Staib L, Stratmann E, Tagare HD, Duncan J, Miller PL: PathMaster: content-based cell image retrieval using automated feature extraction. J Am Med Inf Assoc 7(4):404–415, 2000
Valente F, Costa C, Silva A: Dicoogle, a Pacs featuring profiled content based image retrieval. PLoS ONE 8(5):e61888, 2013
Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H: A comparative study for chest radiograph image retrieval using binary texture and deep learning classification. In 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, 2015, pp 2940–2943
Donner R, Haas S, Burner A, Holzer M, Bischof H, Langs G: Evaluation of fast 2D and 3D medical image retrieval approaches based on image miniatures. In: Müller H, Greenspan H, Syeda-Mahmood T Eds. Medical Content-Based Retrieval for Clinical Decision Support, Vol. 7075. Berlin: Springer Berlin Heidelberg, 2012, pp. 128–138
Kumar A, Kim J, Cai W, Fulham M, Feng D: Content-based medical image retrieval: a survey of applications to multidimensional and multimodality data. Journal of Digital Imaging 26(6):1025–1039, 2013
Le Bozec C, Zapletal E, Jaulent MC, Heudes D, Degoulet P: Towards content-based image retrieval in a HIS-integrated PACS. Proc AMIA Symp:477–481, 2000
Fischer B, Deserno TM, Ott B, Günther RW: Integration of a Research CBIR System with RIS and PACS for Radiological Routine, presented at the Medical Imaging, San Diego, CA, 2008, p. 691914.
Ranjbar S, Whitmire SA, Clark-Swanson KR, Mitchell RJ, Jackson PR, Swanson K: A deep convolutional neural network for annotation of magnetic resonance imaging sequence type. In: In: Society of Imaging Informatics in Medicine, 2019, p. 3
Pizarro R, Assemlal HE, de Nigris D, Elliott C, Antel S, Arnold D, Shmuel A: Using deep learning algorithms to automatically identify the brain MRI contrast: implications for managing large databases. Neuroinformatics 17(1):115–130, 2019
Getting started with pydicom — pydicom 1.1.0 documentation. [Online]. Available: https://pydicom.github.io/pydicom/stable/getting_started.html. [Accessed: 21-Sep-2018].
MongoDB for GIANT Ideas, MongoDB. [Online]. Available: https://www.mongodb.com/index. [Accessed: 21-Sep-2018].
Breiman L: Random forests. Machine Learning 45(1):5–32, 2001
Python Data Analysis Library — pandas: Python Data Analysis Library. [Online]. Available: https://pandas.pydata.org/. [Accessed: 02-Oct-2018].
scikit-learn: machine learning in Python — scikit-learn 0.19.2 documentation. [Online]. Available: http://scikit-learn.org/stable/. [Accessed: 21-Sep-2018].
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
The original list of DICOM attributes was the following:
Image Type, Samples Per Pixel, Photometric Interpretation, Bits Allocated, Bits Stored, High Bit, Scanning Sequence, Sequence Variant, Scan Options, MR Acquisition Type, Repetition Time, Echo Time, Echo Train Length, Inversion Time, Trigger Time, Sequence Name, Angio Flag, Number Of Averages, Imaging Frequency, Imaged Nucleus, Echo Number, Magnetic Field Strength, Spacing Between Slices, Number Of Phase Encoding Steps, Percent Sampling, Percent Phase Field Of View, Pixel Bandwidth, Nominal Interval, Beat Refection Flag, Low RR Value, High RR Value, Intervals Acquired, Intervals Rejected, PVC Rejection, Skip Beats, Heart Rate, Cardiac Number Of Images, Trigger Window, Rate, Reconstruction Diameter, Receive Coil Name, Transmit Coil Name, Acquisition Matrix, In Plane Phase Encoding Direction, Flip Angle, SAR, Variable Flip Angle Flag, DB-Dt, Temporal Position Identifier, Number Of Temporal Positions,Temporal Resolution, Pulse Sequence Name, MR Acquisition Type, Echo Pulse Sequence, Multiple Sin Echo, Multiplanar Excitation, Phase Contrast, Time Of Flight Contrast, Arterial Spin Labeling Contrast, Steady State Pulse Sequence, Echo Planar Pulse Sequence, Saturation Recovery, Spectrally Selected Suppression, Oversampling Phase, Geometry Of K Sapce Traversal, Rectilinear Phase Encode Reordering, Segmented K Space Traversal, Coverage Of K Space, Number Of K Space Trajectories, Pixel Spacing, Slice Thickness, Images In Acquisition, Contrast Bolus Agent.
Rights and permissions
About this article
Cite this article
Gauriau, R., Bridge, C., Chen, L. et al. Using DICOM Metadata for Radiological Image Series Categorization: a Feasibility Study on Large Clinical Brain MRI Datasets. J Digit Imaging 33, 747–762 (2020). https://doi.org/10.1007/s10278-019-00308-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00308-x