Abstract
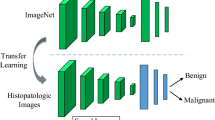
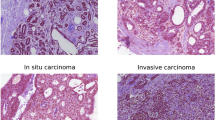
Automatic multi-classification of breast cancer histopathological images has remained one of the top-priority research areas in the field of biomedical informatics, due to the great clinical significance of multi-classification in providing diagnosis and prognosis of breast cancer. In this work, two machine learning approaches are thoroughly explored and compared for the task of automatic magnification-dependent multi-classification on a balanced BreakHis dataset for the detection of breast cancer. The first approach is based on handcrafted features which are extracted using Hu moment, color histogram, and Haralick textures. The extracted features are then utilized to train the conventional classifiers, while the second approach is based on transfer learning where the pre-existing networks (VGG16, VGG19, and ResNet50) are utilized as feature extractor and as a baseline model. The results reveal that the use of pre-trained networks as feature extractor exhibited superior performance in contrast to baseline approach and handcrafted approach for all the magnifications. Moreover, it has been observed that the augmentation plays a pivotal role in further enhancing the classification accuracy. In this context, the VGG16 network with linear SVM provides the highest accuracy that is computed in two forms, (a) patch-based accuracies (93.97% for 40×, 92.92% for 100×, 91.23% for 200×, and 91.79% for 400×); (b) patient-based accuracies (93.25% for 40×, 91.87% for 100×, 91.5% for 200×, and 92.31% for 400×) for the classification of magnification-dependent histopathological images. Additionally, “Fibro-adenoma” (benign) and “Mucous Carcinoma” (malignant) classes have been found to be the most complex classes for the entire magnification factors.












Similar content being viewed by others
References
Breast Cancer. Available at http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/.
Breast Cancer Facts & Figures 2017-2018. Available at https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2017-2018.pdf.
Aubreville M et al.: Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Scientific reports 7:11979, 2017
Wilson ML, Fleming KA, Kuti MA, Looi LM, Lago N, Ru K: Access to pathology and laboratory medicine services: A crucial gap. The Lancet, 2018
Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, Crawford JM, Clark JR, Cantor-Weinberg J, Joshi MG, Cohen MB, Prystowsky MB, Bean SM, Gupta S, Powell SZ, Speights VO Jr, Gross DJ, Black-Schaffer WS: Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Archives of Pathology and Laboratory Medicine 137:1723–1732, 2013
Pöllänen I, Braithwaite B, Haataja K, Ikonen T, Toivanen P: Current analysis approaches and performance needs for whole slide image processing in breast cancer diagnostics. Proc. Embedded Computer Systems: Architectures, Modeling, and Simulation (SAMOS), 2015 International Conference on: City
Veta M, Pluim JP, Van Diest PJ, Viergever MA: Breast cancer histopathology image analysis: A review. IEEE Transactions on Biomedical Engineering 61:1400–1411, 2014
Collins FS, Varmus H: A new initiative on precision medicine. New England Journal of Medicine 372:793–795, 2015
Reardon S: Precision-medicine plan raises hopes: US initiative highlights growing focus on targeted therapies. Nature 517:540–541, 2015
Baba AI, Câtoi C: Comparative oncology: Publishing House of the Romanian Academy Bucharest, 2007
Yn S, Wang Y, Sc C, Wu L, Tsai S: Color-based tumor tissue segmentation for the automated estimation of oral cancer parameters. Microscopy Research and Technique 73:5–13, 2010
Alhindi TJ, Kalra S, Ng KH, Afrin A, Tizhoosh HR: Comparing LBP, HOG and Deep Features for Classification of Histopathology Images. arXiv preprint arXiv:180505837, 2018
Belsare A, Mushrif M, Pangarkar M, Meshram N: Classification of breast cancer histopathology images using texture feature analysis. Proc. TENCON 2015–2015 IEEE Region 10 Conference: City
Rublee E, Rabaud V, Konolige K, Bradski G: ORB: An efficient alternative to SIFT or SURF. Proc. Computer Vision (ICCV), 2011 IEEE international conference on: City
Keskin F, Suhre A, Kose K, Ersahin T, Cetin AE, Cetin-Atalay R: Image classification of human carcinoma cells using complex wavelet-based covariance descriptors. PloS one 8:e52807, 2013
Dheeba J, Singh NA, Selvi ST: Computer-aided detection of breast cancer on mammograms: A swarm intelligence optimized wavelet neural network approach. Journal of biomedical informatics 49:45–52, 2014
Wan S, Huang X, Lee H-C, Fujimoto JG, Zhou C: Spoke-LBP and ring-LBP: New texture features for tissue classification. Proc. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on: City
Zhang Y, Zhang B, Coenen F, Xiao J, Lu W: One-class kernel subspace ensemble for medical image classification. EURASIP Journal on Advances in Signal Processing 2014:17, 2014
Boyd S, El Ghaoui L, Feron E, Balakrishnan V: Linear matrix inequalities in system and control theory: Siam, 1994
Spanhol FA, Oliveira LS, Petitjean C, Heutte L: A dataset for breast cancer histopathological image classification. IEEE Transactions on Biomedical Engineering 63:1455–1462, 2016
Suykens JA, Vandewalle J: Least squares support vector machine classifiers. Neural processing letters 9:293–300, 1999
Breiman L: Random forests. Machine learning 45:5–32, 2001
Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. Proc. Advances in neural information processing systems: City
Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556, 2014
He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Szegedy C, et al.: Going deeper with convolutions. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Zeiler MD, Fergus R: Visualizing and understanding convolutional networks. Proc. European conference on computer vision: City
Lin M, Chen Q, Yan S: Network in network. arXiv preprint arXiv:13124400, 2013
Lakhani P, Gray DL, Pett CR, Nagy P, Shih G: Hello world deep learning in medical imaging. Journal of digital imaging 31:283–289, 2018
LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521:436, 2015
Spanhol FA, Oliveira LS, Petitjean C, Heutte L: Breast cancer histopathological image classification using convolutional neural networks. Proc. Neural Networks (IJCNN), 2016 International Joint Conference on: City
BreakHis Dataset. Available at https://web.inf.ufpr.br/vri/databases/breast-cancer/histopathological-database-breakhis/).
Spanhol FA, Oliveira LS, Cavalin PR, Petitjean C, Heutte L: Deep features for breast cancer histopathological image classification. Proc. Systems, Man, and Cybernetics (SMC), 2017 IEEE International Conference on: City
Araújo T et al.: Classification of breast cancer histology images using convolutional neural networks. PloS one 12:e0177544, 2017
BACH Dataset. Available at https://iciar2018-challenge.grand-challenge.org/Dataset/.
Motlagh NH, et al.: Breast Cancer Histopathological Image Classification: A Deep Learning Approach. bioRxiv:242818, 2018
Sharma S, Mehra R: Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express 4:247–254, 2018
Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S: Breast cancer multi-classification from histopathological images with structured deep learning model. Scientific reports 7:4172, 2017
Vang YS, Chen Z, Xie X: Deep Learning Framework for Multi-class Breast Cancer Histology Image Classification. Proc. International Conference Image Analysis and Recognition: City
Nahid A-A, Kong Y: Histopathological breast-image classification using local and frequency domains by convolutional neural network. Information 9:19, 2018
Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B: Histopathological image analysis: A review. IEEE reviews in biomedical engineering 2:147, 2009
Jeong S: Histogram-based color image retrieval. Psych221/EE362 project report, 2001
Shukla K, Tiwari A, Sharma S: Classification of Histopathological images of Breast Cancerous and Non Cancerous Cells Based on Morphological features. Biomedical and Pharmacology Journal 10:353–366, 2017
Hu M-K: Visual pattern recognition by moment invariants. IRE transactions on information theory 8:179–187, 1962
Lin H, Si J, Abousleman GP: Orthogonal rotation-invariant moments for digital image processing. IEEE Trans Image Processing 17:272–282, 2008
Sonka M, Hlavac V, Boyle R: Image processing, analysis and machine vision London. England: Chapman & Hall Computing:423–431, 1993
Tsai W-H, Chou S-L: Detection of generalized principal axes in rotationally symmetric shapes. Pattern Recognition 24:95–104, 1991
Huang Z, Leng J: Analysis of Hu's moment invariants on image scaling and rotation. Proc. Computer Engineering and Technology (ICCET), 2010 2nd International Conference on: City
Lin W-C, Hays J, Wu C, Kwatra V, Liu Y: A comparison study of four texture synthesis algorithms on regular and near-regular textures. Tech Rep, 2004
Hua B, Fu-Long M, Li-Cheng J: Research on computation of GLCM of image texture [J]. Acta Electronica Sinica 1:155–158, 2006
Haralick RM, Shanmugam K: Textural features for image classification. IEEE Transactions on systems, man, and cybernetics:610–621, 1973
LeCun Y: LeNet-5, convolutional neural networks. URL: http://yannlecuncom/exdb/lenet:20, 2015
Zhang W: Shift-invariant pattern recognition neural network and its optical architecture. Proc. Proceedings of annual conference of the Japan Society of Applied Physics: City
Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
Girshick R, Donahue J, Darrell T, Malik J: Rich feature hierarchies for accurate object detection and semantic segmentation. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
He Y, Zhang X, Sun J: Channel pruning for accelerating very deep neural networks. Proc. International Conference on Computer Vision (ICCV): City
Rabanser S, Shchur O, Günnemann S: Introduction to Tensor Decompositions and their Applications in Machine Learning. arXiv preprint arXiv:171110781, 2017
Lebedev V, Ganin Y, Rakhuba M, Oseledets I, Lempitsky V: Speeding-up convolutional neural networks using fine-tuned cp-decomposition. arXiv preprint arXiv:14126553, 2014
Howard AG, et al.: Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:170404861, 2017
Yamashita R, Nishio M, Do RKG, Togashi K: Convolutional neural networks: an overview and application in radiology. Insights into imaging:1–19, 2018
Chan A, Tuszynski JA: Automatic prediction of tumour malignancy in breast cancer with fractal dimension. Royal Society open science 3:160558, 2016
Nahid A-A, Mehrabi MA: Kong Y: Histopathological Breast Cancer Image Classification by Deep Neural Network Techniques Guided by Local Clustering. BioMed research international 2018, 2018
Veeling BS, Linmans J, Winkens J, Cohen T, Welling M: Rotation equivariant cnns for digital pathology. Proc. International Conference on Medical image computing and computer-assisted intervention: City
Bardou D, Zhang K, Ahmad SM: Classification of Breast Cancer Based on Histology Images Using Convolutional Neural Networks. IEEE Access 6:24680–24693, 2018
Acknowledgements
The authors are immensely thankful to Dr. S. S. Patnaik, director NITTTR, Chandigarh, India, for providing all the necessary facilities and support during the execution of the work. Also, the author Shallu Sharma would like to thank Dr. Sumit Kumar, Associate Professor, Lovely Professional University, Jalandhar, Punjab, India, for every scientific discussion and constant support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, S., Mehra, R. Conventional Machine Learning and Deep Learning Approach for Multi-Classification of Breast Cancer Histopathology Images—a Comparative Insight. J Digit Imaging 33, 632–654 (2020). https://doi.org/10.1007/s10278-019-00307-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00307-y