Abstract
Ottelia, a pantropical genus of aquatic plants belonging to the family Hydrocharitaceae, includes several narrowly distributed taxa in Asia. Although the Asian species have received comparatively more research attention than congeners in other areas, various key taxonomic questions remain unaddressed, especially with regards to apparent cryptic diversity within O. alismoides, a widespread species complex native to Asia, northern Australia and tropical Africa. Here we test taxonomic concepts and evaluate species boundaries using a phylogenetic framework. We sampled five of the seven species of Ottelia in Asia as well as each species endemic to Africa and Australia; multiple samples of O. alismoides were obtained from across Asia. Phylogenetic trees based on five plastid DNA markers and the nuclear ITS region shared almost identical topologies. A Bayesian coalescent method of species delimitation using the multi-locus data set discerned one species in Africa, one in Australia and four in Asia with the highest probability. The results lead us to infer that a population sampled in Thailand represents a hitherto unrecognised cryptic taxon within the widespread species complex, although the apparent lack of unambiguous diagnostic characters currently precludes formal description. Conversely, no molecular evidence for distinguishing O. cordata and O. emersa was obtained, and so the latter is synonymised under the former. Two accessions that exhibit inconsistent positions among our phylogenetic trees may represent cases of chloroplast capture, however incomplete lineage sorting or polyploidy are alternative hypotheses that ought to be tested using other molecular markers.
Similar content being viewed by others
Introduction
Ottelia is a genus of aquatic plants belonging to the family Hydrocharitaceae (Cook and Urmi-König 1984; Cook et al. 1983). The genus has two main centres of diversity, one in tropical Africa (13 species) and the other in Southeast Asia (six species) (Cook and Urmi-König 1984; Cook et al. 1983). In their interfamilial phylogenetic analysis of Alismatidae (Alismatales), Les et al. (1997) included samples of the Asian Ottelia alismoides (L.) Pers. and the African O. ulvifolia (Planch.) Walp. However, significant phylogenetic insights into the subgeneric classification of Ottelia were not gained until Chen et al. (2012) published their molecular phylogenetic analysis of Hydrocharitaceae, in which relationships among five Ottelia species (one accession each of O. acuminata (Gagnep.) Dandy, O. alismoides, O. emersa Z.C. Zhao & R.L. Luo, O. ovalifolia (R.Br.) Rich. and O. sinensis (H. Lév. & Vaniot) H. Lév. ex Dandy) were examined. This study revealed the Australian O. ovalifolia to be sister to the four Asian species. Nevertheless, Ottelia still represents one of the three “hydrocharit genera most in need of more comprehensive study” (Les and Tippery 2013). Of particular interest is the widely distributed O. alismoides, which exhibits extensive morphological variation throughout its range. Molecular systematic studies of a diverse range of other groups containing similarly widespread and morphologically variable taxa, including frogs (Angulo and Icochea 2010; Funket al. 2012), lizards (Oliver et al. 2009), birds (Lohman et al. 2010; Manthey et al. 2011) and crabs (Phiri and Daniels 2016), have uncovered evidence of considerable cryptic speciation.
Cook and Urmi-König (1984) revised the number of Asian species to six, namely, O. acuminata (South China), O. alismoides (widespread in tropical and subtropical Asia and northern Australia), O. balansae (Gagnep.) Dandy (South China and Northern Vietnam), O. cordata (Wall.) Dandy (South China (Hainan), Bangladesh, Cambodia, Myanmar and Thailand), O. mesenterium (Hallier f.) Hartog (Sulawesi) and O. sinensis (South China) (Table 1). A seventh Asian species, O. emersa, was described more recently from Southwest China (Luo and Wang 1987), but He and Sun (1990) rejected the distinctiveness of this entity based on morphological and isozyme evidence, and so synonymised it under O. cordata. Further, He et al. (1990) studied the morphology of O. balansae and O. sinensis and concluded that the two are impossible to distinguish and thus the latter was subsumed under the synonymy of the former. In their treatment of Hydrocharitaceae for the Flora of China, Wang et al. (2010) followed He et al.’s (1990) conclusion in treating O. sinensis as a synonym of O. balansae, but disagreed with He and Sun (1990) and instead accepted both O. cordata and O. emersa as distinct species because “This species [O. emersa] differs from Ottelia cordata by its leaves emersed and not dimorphic, male flowers in each spathe with up to 47–60 flowers, and seeds densely hairy”. Accepted nomenclature according to Govaerts et al. (2018) mostly follows Wang et al. (2010). Ottelia acuminata and O. balansae are morphologically distinct, although identification is sometimes confused if it is ambiguous whether specimens are functionally bisexual or not (Cook and Urmi-König 1984).
The aim of the present study was to assess two taxonomic questions central to an improved understanding of the phylogenetic history of Ottelia: (i) whether undetected cryptic diversity exists within the widespread O. alismoides, and (ii) whether Ottelia cordata and O. emersa are conspecific. To do so, we undertook expanded taxon sampling as compared with the earlier studies, with a particular focus on the species of Asia. Here we apply a phylogenetic species concept to resolve the boundaries within a multi-locus data set comprising both plastid markers (hereinafter ptDNA) and the nuclear ribosomal ITS region (hereinafter nrITS). We utilise our findings to reappraise the taxonomy of the Asian members of the genus.
Materials and methods
Taxon sampling
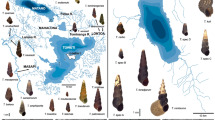
Samples of Ottelia were collected in the field or obtained from herbarium specimens (Fig. 1; Table S1). For specimen identification, we used the taxonomic treatments of Cook et al. (1983) and Cook and Urmi-König (1984), with cross-referencing to the following local floras: den Hartog (1957), Cook (1996), Haynes (2001), Wang et al. (2010) and Tanaka (2015) for Asian species, and Jacobs and McColl (2011) for the Australian species. Hutchinson and Dalziel (1958) was consulted to confirm if our collections from Burkina Faso and Senegal corresponded to O. ulvifolia, the only species recorded in that flora. Identification to infraspecific level within O. acuminata, as defined by Cook and Urmi-König (1984) and Wang et al. (2010), was not conducted because our focus was on inter-specific relationships. Taxa sampled for our study were O. acuminata (three samples, including one from Chen et al. 2012), O. alismoides (20 samples), O. cordata (four samples), O. emersa (a single sample from Chen et al. 2012), O. ovalifolia (one sample), O. sinensis (a single sample from Chen et al. 2012) and O. ulvifolia (four samples) (Fig. 1). Multiple samples of O. alismoides were obtained from across Asia yet no samples were available from northern Australia and tropical Africa. The sample of O. emersa originated from the type locality, which is given as “Guixian (Guiguang), Guangxi, China” (Chen et al. 2012). Blyxa Noronha ex Thouars and Elodea Michx., representing the most and one of the second most closely related genera in the phylogeny of Hydrocharitaceae, respectively, were chosen as outgroup taxa following Tanaka et al. (1997), Les et al. (1997, 2006), Chen et al. (2012), and Les and Tippery (2013), even though Ottelia was found to be non-monophyletic, with some species instead grouping with Blyxa species in the rbcL analyses of Les et al. (1997) and Les and Tippery (2013).
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from silica gel-dried leaf tissue using the CTAB method described in Ito et al. (2010). Five regions of ptDNA (matK, ndhF, rbcL, rpoB and rpoC1) and nrITS were PCR amplified with the following primers: trnK-Hy2F (5′-ACACAGTTTCATACCCAATC) and trnK-Bly3R (5′-CCTTGTTCTGACCATATCGC) for matK; ndhF-F2 (Oxelman et al. 1999) and ndhF-1955R.re (Ito et al. 2017) modified from the primer ndhF-1955R published by Olmstead and Sweere (1994) for ndhF; rbcL-F1F (Wolf et al. 1994) and rbcL-1379R (Little and Barrington 2003) for rbcL; ‘2f’ (5′-ATGCAACGTCAAGCAGTTCC) and ‘4r’ (5′-GATCCCAGCATCACAATTCC) for rpoB (published by Chase et al. 2007); and ‘1f’ (5′-GTGGATACACTTCTTGATAATGG) and ‘3r’ (5′- TGAGAAAACATAAGTAAACGGGC) for rpoC1 (published by Chase et al. 2007); ITS-4 and ITS-5 for nrITS (Baldwin 1992). PCR amplification was conducted using TaKaRa Ex Taq polymerase (TaKaRa Bio, Shiga, Japan). PCR cycling conditions were 94 °C for 60 s; then 30 cycles of 94 °C for 45 s, 52 °C for 30 s, 72 °C for 60 s, with a final extension of 72 °C for 5 min. PCR products were cleaned using ExoSAP-IT purification (GE Healthcare, Piscataway, New Jersey), and then amplified using Big Dye Terminator ver. 3.1 (Applied Biosystems, Foster City, California) using the same primers as those used for the PCR amplifications. DNA sequencing was performed with a 3130xl Genetic Analyzer (Applied Biosystems). Automatic base-calling was checked by eye in Genetyx-Win ver. 3 (Software Development Co., Tokyo, Japan). All sequences generated in the present study have been submitted to the DNA Data Bank of Japan (DDBJ), which is linked to GenBank, and their accession numbers and voucher specimen information are presented in Table S1.
Molecular phylogenetic analysis
Sequences were aligned using Mafft ver. 7.058 (Katoh and Standley 2013) and then inspected manually. Indels were not coded because length variations were either ambiguous (nrITS) or observed only between outgroup and ingroup taxa (matK and ndhF). Analyses were independently performed for ptDNA (matK, ndhF, rbcL, rpoB, and rpoC1) and nrITS data sets respectively to identify possible incongruences between different genomic regions. All 34 ingroup and the two outgroup accessions were included in the ptDNA data set, while 34 ingroup and one outgroup (Blyxa) accessions were included in the nrITS data set to allow accurate alignment of the fast-evolving nrITS region.
Phylogenies were reconstructed using maximum parsimony (MP) in PAUP* ver. 4.0b10 (Swofford 2002), maximum likelihood (ML), and Bayesian inference (BI; Yang and Rannala 1997). In the MP analysis, a heuristic search was performed with 100 random addition replicates and tree-bisection-reconnection (TBR) branch swapping, with the MulTrees option in effect. The MaxTrees option was set at 100,000. Bootstrap analyses (Felsenstein 1985) were performed using 1,000 replicates with TBR branch swapping and simple addition sequences. The MaxTrees option was set at 1,000 to avoid entrapment in local optima.
For the ML analysis, we used the RAxML BlackBox online server (http://phylobench.vital-it.ch/raxml-bb/), which supports GTR-based models of nucleotide substitution (Stamatakis 2008). The maximum likelihood search option was used to find the best-scoring tree after bootstrapping. The gamma model of rate heterogeneity was selected. Statistical support for branches was calculated by rapid bootstrap analyses of 100 replicates (Stamatakis et al. 2008).
BI analyses were conducted with MrBayes ver. 3.2.2 (Ronquist and Huelsenbeck 2003; Ronquist et al. 2012) run on the CIPRES portal (Miller et al. 2010) after the best models had been determined in MrModeltest ver. 3.7 (Nylander 2002); these models were GTR + I+G and HKY + G for ptDNA and nrITS data sets, respectively. Analyses were run for 3,000,000 generations for ptDNA and nrITS data sets, respectively, sampling every 1,000 generations and discarding the first 25% as burn-in. The convergence and effective sampling sizes (ESS) of all parameters were checked in Tracer ver. 1.6 (Rambaut et al. 2014). All trees were visualised using FigTree ver. 1.3.1 (Rambaut 2009). Nodes were recognised as strongly (≥ 90% MP bootstrap support (BS), ≥ 90% ML BS or ≥ 0.95 posterior probability (PP)), moderately (≥ 70% MP BS, ≥ 70% ML BS or ≥ 0.9 PP), or weakly (< 70% MP BS, < 70% ML BS or < 0.9 PP) supported. The data matrices and the MP, RAxML and BI trees were deposited in Treebase (http://purl.org/phylo/treebase/phylows/study/TB2:S20673?x-access-code=ac4d65e4ca13b2a554a1d9522f8d47ee&format=html).
Test of topological incongruence
The Shimodaira Approximately Unbiased (AU) test (Shimodaira 2002) was implemented in PAUP* to test for topological conflict. We used the RAxML alignment in NEXUS format and a file composed of the best trees from each constrained RAxML analysis using the default likelihood settings, with 10,000 bootstrap replicates. The following conflicting nodes discerned upon visual inspection were used as constraints with the other data set: Constraint pt-1, a monophyletic clade of clade VI plus O. alismoides TD5603 and O. alismoides YI1157 in Fig. 2b, and Constraint nr-1, a monophyletic clade of singleton V plus O. alismoides TD5603 and O. alismoides YI1157 in Fig. 2a.
MrBayes trees of Ottelia based on a ptDNA (matK, ndhF, rbcL, rpoB, and rpoC1) and b nrITS data sets. Numbers above or below the branches indicate bootstrap support (BP) calculated in maximum parsimony (MP BS) and maximum likelihood (RAxML BS) analyses and Bayesian prior probabilities (PP). BP < 70% and PP < 0.9 are indicated by hyphens while those of ≥ 95% and ≥ 0.99 are indicated by asterisks. Well-supported clades and singletons are highlighted by grey rectangles with Roman numerals. Two accessions whose position differed between the ptDNA and nrITS trees are enclosed in dashed boxes. Arrows indicate nodes in conflict between the ptDNA and nrITS trees
Species delimitation using STACEY
A Bayesian coalescent method of species delimitation was performed using STACEY (species tree estimation using DNA sequences from multiple loci; Jones 2017), which is an extension of DISSECT (Jones et al. 2015). STACEY was implemented in BEAST ver. 2.4.4 (Bouckaert et al. 2014; Drummond and Rambaut 2007; Drummond et al. 2006).
We ran STACEY using a multilocus data set (ptDNA and nrITS) with all ingroup species; outgroup species were excluded to avoid rate differences and hidden substitutions between ingroup and outgroup species (B. Oxelman, personal communication, November 22, 2016). We performed two independent runs of ten million generations of the MCMC chains, sampling every 1 000 generations. Convergence of the stationary distribution was checked by visual inspection of plotted posterior estimates using Tracer ver. 1.6 (Rambaut et al. 2014). After discarding the first 1 000 trees as burn-in, the samples were summarised in the maximum clade credibility tree using TreeAnnotator ver. 1.6.1 (Drummond and Rambaut 2007) with a posterior probability limit of 0.5 and summarising of mean node heights. The results were visualised using FigTree ver. 1.3.1 (Rambaut 2009).
Results
Molecular phylogeny
The ptDNA data set comprising five genes included 4,611 aligned characters, of which 111 were parsimony-informative. The percentage of missing characters was 14.27% for matK, 50.20% for ndhF, 4.11% for rbcL, 2.17% for rpoB and 3.83% for rpoC1. Analysis of this data set yielded the imposed limit of 100 000 MP trees (tree length = 395 steps; consistency index = 0.90; retention index = 0.92). The strict-consensus MP tree, the RAxML tree, and the MrBayes BI 50% consensus tree showed no incongruent phylogenetic relationships; thus, only the MrBayes tree is presented here (Fig. 2a).
It is ambiguous which species of Ottelia diverged first. In the RAxML analysis, O. ovalifolia (singleton II) is sister to all remaining species (58% RAxML BS), whereas in the MrBayes tree, O. ulvifolia (clade I) is sister to the rest (0.51 PP); neither relationship receives statistical support in the MP analysis. The core group excluding these two species is supported in both the RAxML (84% RAxML BS) and MrBayes (1.0 PP) analyses, and is resolved as comprising two distinct lineages: one containing clade III (O. cordata-O. emersa), singleton V (O. alismoides YI1707), O. alismoides TD5603 and O. alismoides YI1157 (61% RAxML BS; 1.0 PP), and the other containing clade IV (O. acuminata–O. sinensis) and clade VI (O. alismoides) (55% RAxML BS; 0.73 PP). However, neither of these lineages is supported in the MP tree.
The nrITS data set included 845 aligned characters, of which 216 were parsimony-informative. The percentage of missing characters was 12.97%. Analysis of this data set yielded the imposed limit of 100,000 MP trees (tree length = 538 steps; consistency index = 0.83; retention index = 0.93). The strict-consensus MP tree, the RAxML tree and the MrBayes BI 50% consensus tree showed no incongruent phylogenetic relationships; thus, only the MrBayes tree is presented here (Fig. 2b).
It is again ambiguous which species of Ottelia diverged first. In the RAxML and MrBayes analyses, O. ovalifolia (singleton II) is sister to all other taxa (63% RAxML BS; 0.75 PP), whereas in the MP tree O. ulvifolia (clade I) is sister to them (69% MP BS). The core group excluding these two species is strongly supported (98% MP BS; 100% RAxML BS; 1.0 PP) and clearly divided into two lineages: one comprising clade III (O. cordata–O. emersa) and clade IV (O. acuminata–O. sinensis) (63% MP BS; 88% RAxML BS; 0.97 PP), and the other comprising singleton V (O. alismoides YI1707), clade VI (O. alismoides), O. alismoides TD5603 and O. alismoides YI1157 (90% MP BS; 95% RAxML BS; 1.0 PP).
Topological incongruence
We observed two cases of hard incongruence (≥ 70% BS; Johnson and Soltis 1998) between ptDNA and nrITS trees, both of which were attributed to two accessions, O. alismoides TD5603 and O. alismoides YI1157: the two were positioned next to singleton V in the ptDNA data set, but were grouped in clade VI in the nrITS data set (arrows in Fig. 2). Constraining these conflicting nodes onto the other data set retrieved trees with maximum likelihood scores that were significantly worse than those derived from an unconstrained search, according to the AU test (P < 0.05 in both analyses; Tables S2, S3).
Species delimitation using STACEY
SpeciesDelimitationAnalyser generated 265 clusterings from the MCMC runs. The highest posterior probability was 0.33 PP for a classification with six species or minimal clusters, i.e. comprising four clades and two singletons (Fig. 3). The similarity matrix revealed that the individuals within the four clades had less than 0.07 PP belonging to a different cluster or a singleton (data not shown). While the similarity score among the 17 O. alismoides samples was 0. 9 858 739 on average, the score between these 17 samples and the single Ottelia sp. sample averaged 0. 0997812 (Table S4). Similarly, the average similarity score among the three samples of O. acuminata was 0. 9790857, whereas the score between the three O. acuminata samples and one O. sinensis sample was 0. 9255969 (Table S4). Further, the average similarity score among the four O. cordata samples was 0. 9544697, but the score between these four samples and the single O. emersa sample was 0. 9669628 (Table S4).
Maximum sum of clade credibility SMC-tree based on multi-locus (plastid DNA, nuclear ITS) data set of Ottelia from BEAST 2 analysis and similarity matrix from STACEY analysis. Posterior probabilities from BEAST 2 are given for major clades on the branches. The squares in the matrix represent posterior probabilities (white = 0, black = 1) for pairs of individuals belonged to the same cluster. Roman numerals on the clades of the SMC-tree, correspond to those in Fig. 2, representing the six minimal clusters or species of Ottelia delimited in STACEY analysis. The similarity scores of the two accessions that exhibited conflicting placement in Fig. 2 are indicated with dotted lines
Discussion
The present study examined intergeneric relationships within Ottelia using a phylogenetic approach. A Bayesian coalescent method of species delimitation using the multi-locus data set discerned six species in our sample set. Here we discuss the implications of these findings in terms of the taxonomy of Ottelia species drawing on the phylogenetic species concept and with a particular focus on traditional morphological characters to distinguish species.
Cryptic diversity within Ottelia alismoides
Multiple molecular systematic studies have uncovered cryptic diversity within widespread taxa belonging to various life forms. In the present study, we sought to re-evaluate the status of the geographically widespread and morphologically variable Ottelia alismoides (Cook and Urmi-König 1984), which is conventionally diagnosed by its “spathe with 3 or more wings” (Cook and Urmi-König 1984; Cook et al. 1983), through phylogenetic analysis of multiple samples collected throughout its range in Asia and encompassing much of the morphological variation reviewed by Cook and Urmi-König (1984). Ottelia alismoides is, in all of our molecular phylogenetic analyses, divided into two lineages (Fig. 2), both of which are also recovered in species delimitation analysis (with an average similarity score of < 0.1) as belonging to the same species or minimal clusters (Fig. 3; Table S4). We therefore infer that O. alismoides sensu Cook and Urmi-König (1984) contains at least one cryptic species, in addition to O. alismoides sensu stricto (hereinafter referred to as O. alismoides s.s.). This cryptic species is represented in our study by the single accession YI1707 from Thailand.
Cook and Urmi-König (1984) summarised the extensive morphological variation in O. alismoides in terms of discrepancies in leaf outline (narrowly elliptic to very widely ovate), spathe morphology (urceolate, ellipsoidal, or cylindrical), flower colour (white, pink, bluish, violet, or light purple), and the number of stamens (3–12) and carpels (3–10). We confirmed much of this morphological variation among the 20 accessions of O. alismoides sensu Cook and Urmi-König (1984) included in our phylogenetic analyses, but nevertheless found no clear-cut difference between O. alismoides s.s. and O. alismoides YI1707. Given the wide karyotype variation that has been reported within O. alismoides, with diploid numbers ranging from 2n = 22 to 2n = 132 (Cook and Urmi-König 1984), it is possible that the cryptic taxon detected here may be distinguishable on the basis of chromosome number, but this character was not assessed in our samples. For the time being, we refrain from making any firm conclusion on the taxonomic status of O. alismoides until further material belonging to the same operational taxonomic unit as O. alismoides YI1707 becomes available.
Conspecific nature of Ottelia cordata and O. emersa
The present study aimed to test two alternative hypotheses pertaining to the status of Ottelia cordata and O. emersa in tropical Asia: He and Sun (1990) rejected the distinctiveness of the two species based on morphological and isozyme evidence, whereas Wang et al. (2010) and WCSP (2018) accepted both. The present study included four accessions of O. cordata and a single sample of O. emersa from the type locality in Guangxi, South China (Chen et al. 2012). The results of our phylogenetic analyses reveal O. emersa to be nested within O. cordata (with an average similarity score of 0. 9669628; Table S4), suggesting that the two are conspecific (Figs. 2, 3). Given that O. cordata is the earlier name, it takes nomenclatural priority over O. emersa (McNeil et al. 2012: Art. 11.3). The morphological differences between the two entities documented by Wang et al. (2010) (i.e. leaves emersed, submerged or floating, 10–30 or 47–60 male flowers per spathe and seeds densely hairy or smooth), should accordingly be considered infraspecific variation, and thus the description of O. cordata is here revised (see Taxonomic treatment).
Are Ottelia acuminata and O. sinensis distinct?
Ottelia acuminata is a dioecious species described from southwest China (Cook and Urmi-König 1984). It has since been recorded also from southeast China (Wang et al. 2010). In contrast, O. sinensis is a monoecious species known to occur in southwest China (Cook and Urmi-König 1984) and in northern Vietnam under the synonym O. balansae. The present study indicates that the single accession from Guangxi, southeast China, identified as O. sinensis by Chen et al. (2012), is phylogenetically indistinguishable from the three accessions of O. acuminata (with an average similarity score of 0.93; Table S4), possibly suggesting that the two are conspecific. However, because the locality in Guangxi has been reported to harbour both O. acuminata and O. sinensis (Cook and Urmi-König 1984; Wang et al. 2010), we cannot rule out the possibility that Chen et al. (2012) misidentified O. acuminata as O. sinensis. For the time being, we refrain from making any firm conclusion on the taxonomic status of either species until further material of O. sinensis becomes available, ideally from northern Vietnam where O. sinensis has been reported (under the synonym O. balansae), but where O. acuminata is not known to be distributed.
Evidence of reticulate evolution in Ottelia?
Two accessions of Ottelia alismoides from Myanmar and India (TD5603, YI1757) occupy inconsistent positions in the ptDNA and nrITS trees (Fig. 2; Tables S2, S3). Cases of such topological incongruence are often reported in phylogenetic studies (reviewed by Degnan and Rosenberg 2009; Wendel and Doyle 1998). Among the known causes, which include the particular genetic marker selected for phylogenetic reconstruction and incomplete lineage sorting (Wendel and Doyle 1998), chloroplast capture may best explain the phenomenon in Ottelia because the two accessions have unique ptDNA haplotypes but share nrITS ribotypes with O. alismoides, and also lack unique, heterogeneous nrITS (Fig. 2). It is noteworthy that the two accessions may represent independent taxa of hybrid origin, as in the recently reported case involving Nymphoides montana Aston (Menyanthaceae; Tippery and Les 2011), but this scenario is less likely because the specimens exhibited no distinct morphological differences. Additional material representing these entities from the two countries will be subjected to further phylogenetic and taxonomic analysis, in which alternative, more discriminatory approaches, such as low-copy nuclear DNA markers and AFLP, will be applied in order to provide greater clarity on the cause of this ambiguous finding.
Taxonomic treatment
Ottelia cordata (Wall.) Dandy, J. Bot. (London), 72: 137, 1934.
≡ Boottia cordata Wall., Pl. Asiat. Rariores, 1: 52, t. 65, 15 July 1830.—TYPE: BURMA. “ripae Irawaddi prope Avam” (Ava). Lectotype (designated by Cook and Urmi-König in Aquat. Bot. 20: 146. 1984): t. 65 in Wall., Pl. Asiat. Rariores, 1. 15 July 1830 (non vidi photo!).
= Ottelia emersa Z.C. Zhao et R.L. Luo, J. Wuhan Bot. Res., 5: 339, f. 1, November 1987. —TYPE: CHINA. Guangxi, Guixian county, in ponds and canals. Lectotype (designated by He and Sun in Aquat. Bot. 36: 397. 1990): 8 Oct. 1980, Z.C. Zhao 0595 (WH non vidi); paratype (designated herein): 22 Sep. 1986, R.L. Luo 030 (WH non vidi).
Diagnostic Features—Specimens having only emmersed leaves, densely hairy seeds, and 47–60 male flowers in the male spathe that were identified by Wang et al. (2010) as O. emersa are here treated as morphological variation within O. cordata.
Distribution and Habitat—Cambodia, China (Guangxi, Hainan), Myanmar, Thailand, Vietnam.
Notes—Two specimens were cited in the protologue of Ottelia emersa by Luo and Wang (1987). He and Sun (1990) apparently chose the older one (Z.C. Zhao 0595) as a lectotype; thus, the other specimen (R.L. Luo 030) is herein selected as a paratype, which He and Sun (1990) incorrectly treated as an “isotype”.
IUCN conservation assessment—More than ten populations are known to occur in China (Guangxi, Hainan), Myanmar, Thailand, and Vietnam, including those confirmed in the course of the present study, with a total population size in excess of 1,000 individuals. Plants are known to set fruit and recruit freely at most of these sites. Although wetlands are in general threatened by land conversion and other modifications to natural drainage patterns throughout this region, there is at present no evidence that this species has undergone decline as a result. Therefore, we follow the current conservation assessment of Least Concern (LC) prepared by Juffe-Bignoli (2011).
References
Angulo A, Icochea J (2010) Cryptic species complexes, widespread species and conservation: lessons from Amazonian frogs of the Leptodactylus marmoratus group (Anura: Leptodactylidae). Syst Biodiv 8:357–370
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol 1:3–16
Bouckaert RR, Heled J, Kühnert D, Vaughan TG, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñan S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TE, Kelly L, Wilkinson M (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56:295–299
Chen LY, Chen JM, Gituru RW, Wang QF (2012) Generic phylogeny, historical biogeography and character evolution of the cosmopolitan aquatic plant family Hydrocharitaceae. BMC Evol Biol 12:30
Cook CDK (1996) Aquatic and wetland plants of India. Oxford University, Oxford
Cook CDK, Urmi-König K (1984) A revision of the genus Ottelia (Hydrocharitaceae). 2. The species of Eurasia, Australasia and America. Aquat Bot 20:131–177
Cook CDK, Symoens J-J, Urmi-König K (1983) A revision of the genus Ottelia (Hydrocharitaceae). 1. Generic considerations. Aquat Bot 18:263–274
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24:332–340
den Hartog C (1957) Hydrocharitaceae. In: van Steenis CGGJ (ed) Flora Malesiana, ser. 1, vol 5. Noordhoff-Kollf. Djakarta, pp 381 − 413
Drummond AJ, Rambaut A (2007) BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214–222
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Funk WC, Caminer M, Ron SR (2012) High levels of cryptic species diversity uncovered in Amazonian frogs. Proc R Soc B 279:1806–1814
Govaerts R (2018) World checklist of Hydrocharitaceae. Facilitated by the royal botanic gardens, Kew. Published online at http://apps.kew.org/wcsp/. Retrieved 22 December 2018
Haynes RR (2001) Hydrocharitaceae. In: Santisuk T, Larsen K (eds) Flora of Thailand, vol 7. The Forest Herbarium. Royal Forest Department, Bangkok, pp 365–382
He J-B, Sun X-Z (1990) Ottelia emersa Z.C. Zhao et R.L. Luo (Hydrocharitaceae)—a Synonym of O. cordata (Wallich) Dandy. Aquat Bot 36:395–398
He J-B, Sun X-Z, Wang H-Q (1990) Taxonomy of the bisexual species of Ottelia in China. Aquat Bot 36:389–393
Hutchinson J, Dalziel JM (1958) Flora of west tropical Africa vol. I. part 2 (2nd edn revised by Keay RWJ). Royal Botanic Gardens, Kew, pp 1662–1664
Ito Y, Ohi-Toma T, Murata J, Tanaka N (2010) Hybridization and polyploidy of an aquatic plant, Ruppia (Ruppiaceae), inferred from plastid and nuclear DNA phylogenies. Am J Bot 97:1156–1167
Ito Y, Tanaka N, Albach DC, Barfod AS, Oxelman B, Muasya AM (2017) Molecular phylogeny of the cosmopolitan aquatic plant genus Limosella (Scrophulariaceae) with a particular focus on the origin of the Australasian L. curdieana. J Plant Res 140:107–116
Jacobs SWL, McColl KA (2011) Hydrocharitaceae. In: Wilson AJG (ed) Flora of Australia, vol 39. ABRS/CSIRO, Melbourne, pp 14–44
Johnson LA, Soltis DE (1998) Assessing congruence: empirical examples from molecular data. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematic of plants II: DNA sequencing. Kluwer Academic Publisher, Norwell, pp 265–296
Jones GL (2017) Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J Math Biol 74:447–467
Jones GL, Aydin Z, Oxelman B (2015) DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 31:991–998
Juffe-Bignoli, D (2011) Ottelia cordata. The IUCN red list of threatened species 2011: e.T194035A8879309. http://dx.doi.org/10.2305/IUCN.UK.2011-2.RLTS.T194035A8879309.en. Downloaded on 09 January 2019
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Les DH, Tippery NP (2013) In time and with water. the systematics of alismatid monocotyledons. In: Wilkin P, Mayo SJ (eds) Early events in monocot evolution. Cambridge University Press, Cambridge, pp 119–164
Les DH, Cleland MA, Waycott M (1997) Phylogenetic studies in Alismatidae, II: evolution of marine angiosperms (‘seagrasses’) and hydrophily. Syst Bot 22:443–463
Les DH, Moody ML, Soros C (2006) A reappraisal of phylogenetic relationships in the monocotyledon family Hydrocharitaceae. In: Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG (eds) Monocots: comparative biology and evolution: excluding Poales. Rancho Santa Ana Botanical Garden, Claremont, pp 211–230
Little DP, Barrington DS (2003) Major evolutionary events in the origin and diversification of the fern genus Polystichum (Dryopteridaceae). Am J Bot 90:508–514
Lohman DJ, Ingram KK, Prawiradilaga DM, Winker K, Sheldon FH, Moyle RG, Ng PKL, Ong PS, Wang LK, Braile TM, Astuti D, Meier R (2010) Cryptic diversity in “widespread” southeast Asian bird species suggests that Philippine avian endemism is gravely underestimated. Biol Conserv 143:1885–1890
Luo RL, Wang HQ (1987) A new species of Ottelia (Hydrocharitaceae) and its karyotype. J Wuhan Bot Res 5:339–342
Manthey JD, Klicka J, Spellman GM (2011) Cryptic diversity in a widespread North American songbird: phylogeography of the Brown Creeper (Certhia americana). Mol Phylogenet Evol 58:502–512
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prud’homme Van Reine WF, Smith GF, Wiersema JH, Turland NJ (eds) (2012) International code of nomenclature for algae, fungi and plants (Melbourne Code): Adopted by the Eighteenth International Botanical Congress, Melbourne, Australia, July 2011. Regnum Vegetabile 154. Koeltz Scientific Books, Königstein
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), 14 Nov. 2010, New Orleans, pp 1–8. http://www.phylo.org/sub_sections/portal/cite.php
Nylander JAA (2002) MrModeltest. Ver. 1.0. Program distributed by the author. Department of Systematic Zoology, Uppsala University, Uppsala. Retrieved from http://www.ebc.uu.se/systzoo/staff/nylander.html
Oliver PM, Adams M, Lee MSY, Hutchinson MN, Doughty P (2009) Cryptic diversity in vertebrates: molecular data double estimates of species diversity in a radiation of Australian lizards (Diplodactylus, Gekkota). Proc R Soc B 276:2001–2007
Olmstead RG, Sweere JA (1994) Combining data in phylogenetic systematics: an empirical approach using three molecular data sets in the Solanaceae. Syst Biol 43:467–481
Oxelman B, Backlund M, Bremer B (1999) Relationships of the Buddlejaceae s. l. inferred from chloroplast rbcL and ndhF sequences. Syst Bot 24:164–182
Phiri EE, Daniels SR (2016) Multilocus coalescent species delimitation reveals widespread cryptic differentiation among Drakensberg mountain-living freshwater crabs (Decapoda: Potamonautes). Invertebr Syst 30:60–74
Rambaut A (2009) FigTree ver. 1.3.1: Tree Figure Drawing Tool. Retrieved from http://tree.bio.ed.ac.uk/software/figtree/
Rambaut A, Suchard MA, Xie W, Drummond AJ (2014) Tracer. ver. 1.6. Retrieved from http://beast.bio.ed.ac.uk/Tracer
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771
Swofford DL (2002) PAUP: Phylogenetic analysis using parsimony (and other methods). ver. 40b10. Sinauer Associates, Sunderland
Tanaka N (2015) Hydrocharitaceae. In: Ohashi H, Kadota Y, Murata J, Yonekura K, Kihara H (eds) Wild flowers of Japan, vol 1. Heibonsha, Tokyo, pp 118–125 (in Japanese)
Tanaka N, Setoguchi H, Murata J (1997) Phylogeny of the family Hydrocharitaceae inferred from rbcL and matK gene sequence data. J Plant Res 110:329–337
Tippery NP, Les DH (2011) Evidence for the hybrid origin of Nymphoides montana Aston (Menyanthaceae). Telopea 13:285–294
Wang QF, Guo YH, Haynes RR, Hellquist CB (2010) Hydrocharitaceae. In: Wu C-Y, Raven PH, Hong D-Y (eds) Flora of China, vol 23. Science Press. Beijing & Missouri Botanical Garden Press, St. Louis, pp 91–102
Wendel JF, Doyle JJ (1998) Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants II. Kluwer Academic Publishing, Boston, pp 265–296
Wolf PG, Soltis PS, Soltis DE (1994) Phylogenetic relationships of Dennstaedtioid ferns: evidence from rbcL sequences. Mol Phylogenet Evol 3:383–392
Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol 14:717–724
Acknowledgements
The authors thank H. Wang (HITBC) and E. Liu (KUN) for arranging loans from their institutions and/or for hospitality during the authors’ recent visits; R. Pooma, S. Saengrit, N. Suphuntee, K. Chayamarit (BKF), K. Shuto (Fukushima), S.R. Yadav (Shivaji), H. Murata (Osaka), T. Sugawara (MAK), Nb. Tanaka (TNS), J. Murata, T. Ohi-Toma (TI), A. Naiki, Y. Saito (Okinawa) for assistance in the field; and C. Ishii (Tsukuba) for help with DNA sequencing. This research was supported by Chinese Academy of Sciences (CAS) President’s International Fellowship Initiative (PIFI) Grant no. 2015PB022 to YI and by the ‘Integrated analysis of natural history collections for conservation of highly endangered species’ programme under the National Museum of Nature and Science, Japan to NT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ito, Y., Tanaka, N., Barfod, A.S. et al. Molecular phylogenetic species delimitation in the aquatic genus Ottelia (Hydrocharitaceae) reveals cryptic diversity within a widespread species. J Plant Res 132, 335–344 (2019). https://doi.org/10.1007/s10265-019-01109-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-019-01109-7