Abstract
Plant cytokinesis is achieved by formation of cell plates in the phragmoplast, a plant-specific cytokinetic apparatus, which consists of microtubules (MTs) and microfilaments. During cytokinesis, the cell plate is expanded centrifugally outward from the inside of cells in a process that is supported by dynamic turnover of MTs. M-phase-specific kinesin NACK1, which comprises the motor domain at the amino-terminal half to move on MT bundles and the stalk region in the carboxyl-terminal half, is a key player in the process of MT turnover. That is, the specific region in the stalk binds the MAP kinase kinase kinase to activate the whole MAP kinase cascade, which stimulates depolymerization of MTs for the MT turnover. The stalk is also responsible for recruiting the activated kinase cascade to the mid-zone of the phragmoplast, which corresponds to the cell-plate formation site. It should be crucial to uncover roles of the NACK1 kinesin stalk as well as the motor domain in the formation of cell plates in order to understand the mechanisms of cell plate formation. Using dissected Arabidopsis NACK1 (AtNACK1/HINKEL) molecules and AtNACK1-fused GFP, we showed that the C-terminal tail of the stalk in addition to the motor domain is critical for its proper localization to the site of cell plate formation in the phragmoplast, probably by affecting its motility activity.
Similar content being viewed by others
Introduction
Cytokinesis is the process whereby one cell physically separates into two at the end of the cell cycle, which is essential for the growth and development of multicellular organisms. If cytokinesis fails to occur in animals, multinucleate cells, heteroploid cells, and aberrantly amplified centrosomes are formed, which are often related to tumorigenicity and apoptosis of cells (Normand and King 2010). In plants, defects in cytokinesis cause the generation of multinucleate cells and accumulation of stubs containing premature cell plates in aberrantly enlarged cells, which results in dwarfism of plant bodies and organs (Nishihama and Machida 2001; Nishihama et al. 2002; Sazuka et al. 2005; Strompen et al. 2002).
Plant cytokinesis is achieved by the formation of a cell plate in the characteristic cytokinetic machinery of plant cells called the phragmoplast, which is mainly composed of aligned microtubules (MTs) and actin filaments (Müller et al. 2009; Nishihama and Machida 2001; Otegui et al. 2005). Formation of the phragmoplast starts at a certain time in anaphase of the late M phase between two separating daughter nuclei in the internal region of the cytoplasm (Nishihama and Machida 2001; Otegui et al. 2005). As cytokinesis proceeds in a cell, bundles of phragmoplast MTs expand centrifugally from the interior to the periphery of the cell, driving the centrifugal development of a cell plate behind the expanding phragmoplast MTs. In the newly synthesized cell plate, a new cell wall is constructed within the lumens of membranous tubules that are formed by the fusion of membrane vesicles transported by membrane trafficking systems with MTs (Jürgens 2005; McMichael and Bednarek 2013; Van Damme and Geelen 2008; Woollard and Moore 2008). Although some vesicle trafficking-related components such as KNOLLE, Rab-A GTPases, and PATL1 have been identified (Chow et al. 2008; Lauber et al. 1997; Peterman et al. 2004; Waizenegger et al. 2000), the molecular mechanisms for the trafficking involving these factors in the formation of cell plates have yet to be determined.
The centrifugal expansion of the phragmoplast is supported by a dynamic turnover of MTs involving the depolymerization of MTs inside of the bundle, while MTs are newly generated (elongation) outside of the bundle (Murata et al. 2013; Nakaoka et al. 2012; Smertenko et al. 2011; Yasuhara et al. 1993). Several MT-associated proteins (MAPs) including MAP65, MOR1/GEM1 (XMAP215), and kinesin-like proteins stimulate the turnover of MTs (Müller et al. 2004; Nishihama et al. 2002; Sasabe et al. 2006; Strompen et al. 2002; Twell et al. 2002).
Expansion of the phragmoplast in tobacco BY-2 cells is positively regulated by the mitogen-activated protein (MAP) kinase cascade, all the components of which localize to the equatorial zone of the phragmoplast corresponding to the site of cell plate formation (Ishikawa et al. 2002; Nishihama et al. 2001; Soyano et al. 2003). Activity of the cascade is controlled by cytokinesis-specific kinesins, which are designated NACK1 and NACK2 in tobacco cells. NACK1 protein is located in the fraction that is purified biochemically from phragmoplasts by following the microtubule-based motility activity, suggesting that NACK1 may have a phragmoplast-associated motility activity (Nishihama et al. 2002). Both kinesins localize to the equatorial zone of the phragmoplast during cytokinesis and directly bind to the NPK1 MAP kinase kinase kinase (MAPKKK) to increase NPK1 activity at late M phase during cytokinesis (Nishihama et al. 2002), and this process is inhibited by phosphorylation of NACK1 and NPK1 by cyclin-dependent kinase (CDKs) before metaphase (Sasabe et al. 2011a). These kinesins have the MT-based motor domains at the amino-termini and the stalk regions in the carboxyl-terminal halves that are rich in several predicted coiled-coils structures, one of which provide binding sites for NPK1 MAPKKK, and they also consistently localize to the equatorial region of the phragmoplast during its expansion. Thus, NACK1 kinesin recruits NPK1 MAPKKK, which binds NQK1 MAP kinase kinase (MAPKK) and NRK1 MAP kinase (MAPK) to the phragmoplast equator, in addition it activates NPK1 and the whole kinase cascade (Nishihama et al. 2002). On the basis of the above results, the NACK-PQR MAPK pathway including these components is proposed to positively facilitate expansion of the phragmoplast (Soyano et al. 2003). Activated NRK1 MAPK phosphorylates MAP65 to reduce its activity of bundling MTs, which increases the rate of the MT turnover to allow phragmoplast MT arrays to expand centrifugally (Sasabe et al. 2006).
Genetic analyses have shown that a similar pathway is also conserved in Arabidopsis thaliana (Kosetsu et al. 2010; Sasabe et al. 2011b; Takahashi et al. 2010). AtNACK1/HINKEL (HIK), an Arabidopsis homolog of NACK1, appears to be a key regulator of the progression of cytokinesis, because knockout and knockdown mutants of AtNACK1/HIK and/or AtNACK2/STUD (STD)/TETRASPORE (TES) show defects in cell plate structures and gametophytic lethality in Arabidopsis plants (Tanaka et al. 2004). The myc-tagged AtNACK1/HIK protein is also localized to the phragmoplast equator (Krupnova et al. 2013). In addition to Arabidopsis and rice (Sazuka et al. 2005), the genomes of all multicellular plants so-far examined encode for proteins that are homologous to NACK1 (Miki et al. 2014), suggesting a ubiquitous role of these homologues in cytokinesis. The cellular biological information of AtNACK1/HIK in Arabidopsis, however, is still limited. In the present study, we took advantage of the tobacco cell line BY-2, which is considered to be highly proliferative, to investigate the molecular and cellular bases for the characteristics of AtNACK1/HIK. We have investigated which regions of AtNACK1 are required for its localization to the equatorial zone of the phragmoplast corresponding to the site of cell plate formation. The results in the present study show that the C-terminal region of the stalk, as well as the motor domain, is necessary for the proper localization of AtNACK1/HIK to the phragmoplast equatorial zone.
Materials and methods
DNA constructs
The deletion constructs were made by PCR and overlap PCR. Each fragment of AtNACK1 cDNA was amplified by PCR with specific primers and/or overlap primers (sequences of these primers will be sent on request) and cloned into the pENTR1A vector (Invitrogen, Carlsbad, CA). To express GFP fusions in BY-2 cells, the fragments of AtNACK1 cDNAs in pENTR were subcloned into the binary vector pGWB452 (Nakagawa et al. 2007) by using Gateway® LR Clonase II (Invitrogen). All of these constructs were N-terminally GFP-fused. To express N-terminal GFP fusion in cells by estradiol, we introduced the sGFP-fused Gateway® cassette from the pGWB6 vector (Nakagawa et al. 2007) into the pER8 vector that contains the estradiol-inducible promoter sequence (XVE sequence) (Zuo et al. 2000), which yielded the pER8GW6 vector. The full length cDNA fragment of AtNACK1 was also subcloned into the binary vector pER8GW6 by the Gateway system. To express the C-terminal GFP fusion in cells with estradiol, the AtNACK1-GFP fragment was amplified by PCR with the C-terminally GFP-fused AtNACK1 sequence cloned in pGWB451 (Nakagawa et al. 2007) as a template, and cloned into pER8 vector. The 6.1-kb DNA fragment covering the AtNACK1 locus from a position 1.3 kb 5′-upstream of the initiation codon to a position 0.75 kb 3′-downstream of the termination codon (Sasabe et al. 2011a) was used as a template to make a construct which expressed a GFP-fusion protein under the original promoter of AtNACK1. The 5.4-kb DNA fragments, excluding the 3′ UTR of AtNACK1, were amplified by specific primers and cloned into the pCR8 vector (Invitrogen). This fragment was sequentially subcloned into the binary vector pGWB504 (Nakagawa et al. 2007) to fuse sGFP to the C-terminal of AtNACK1. The sequences of all constructs were confirmed by sequencing analysis.
Plant materials, transformations, and selection of transgenic lines
Tobacco BY-2 cells were maintained in suspension culture at 26 °C in darkness with weekly subculture in modified Linsmaier and Skoog medium. Arabidopsis thaliana plants (Col-0) were grown with a 16-h photoperiod at 22 °C either in soil or on Murashige and Skoog (MS) plates that contained 0.8 or 1.5 % agar. Each construct was transformed into BY-2 cells and/or wild-type Col-0 plants by using the Agrobacterium tumefaciens-mediated method (An 1985; Clough and Bent 1998). More than 10 lines of transformants including each construct of BY-2 were used for observation of subcellular localization, and the offspring of seven independent T1 plants were used for subsequent analysis. The BY-2 cells transformed with transgenes under the control of an estradiol-inducible promoter were cultured in liquid medium containing 0.1 µM 17-β-estradiol (Sigma-Aldrich, St Louis, MO, USA) for 16 h to express GFP fusion.
Microscopy
BY-2 calli were suspended in liquid medium and used for observation. GFP fluorescence was detected with an AXIO Imager (Carl Zeiss, Oberkochen, Germany) and a confocal microscope, FV1000 (Olympus, Tokyo, Japan). Transgenic plants for microscopic observation were grown on MS plates containing 1.5 % agar. Plates were placed vertically in growth chambers for 7–14 days. To observe the dividing cells of root-tips in the 7-day-old transgenic plants, a 10 µM solution of FM4–64 (Invitrogen) was applied dropwise onto the root-tips, which were then incubated for 10 min. Fluorescence due to GFP and FM4–64 was sequentially recorded by confocal microscopy (LSM780; Carl Zeiss).
Phenotypic analysis
Leaves from 14-day-old plants grown on the MS plates were cleared, basically, as described by Ishikawa et al. (2003). Leaves were fixed overnight at room temperature in a mixture of ethanol and acetic acid (9:1, v/v). After rehydration in a graded ethanol series (90, 70, 50, and 30 % for 20 min each), they were cleared in a solution of chloral hydrate (trichloroacetaldehyde monohydrate, 80 g; glycerol, 10 ml; distilled water, 20 ml). Cleared leaves were observed with by Nomarski differential interference microscopy (AxioPlan2; Carl Zeiss).
Results
GFP-AtNACK1 localizes to the site of cell plate formation, expands to the parental cell walls, and disappears in living tobacco BY-2 cells
As is the case for tobacco NACK1 (Nishihama et al. 2002), in silico analysis by means of the InterPro (http://www.ebi.ac.uk/interpro/) and COILS programs (Lupas et al. 1991) predicts with high probability the presence of a putative motor domain (amino acids 1–360) and several coiled-coil structures in the carboxyl-terminal (C-terminal) half, which is called the stalk region in NACK1, of AtNACK1/HIK (Fig. 1a, residues 361–974). In addition to the highly probable coiled-coil structures, a single coiled-coil region is predicted with lower probability in the C-terminal tail (Fig. 1a, residues 764–974).
Domain and motif organizations of AtNACK1. a The probability of coiled-coil formation plotted against the amino acid number for AtNACK1 by using the COIL program and a 28-amino acid prediction window (Lupas et al. 1991). b Schematic representation of wild-type and deletion mutant constructs of AtNACK1/HIK. Predicted domain organization and relevant amino-acid positions for the AtNACK1 protein sequence are indicated in the wild-type schematic diagram. The coding region of the stalk (ST) is divided into three parts (ST-A, ST-B, and ST-C), which were fused to the region corresponding to the predicted motor domain (MD) and designated as AtNACK1:MD-A, MD-AB, MD-AC, and MD-BC. The coding sequence corresponding to each construct was fused to the C-terminus of the sequence of G3GFP or sGFP
We made the XVE::GFP-AtNACK1 construct, in which the coding sequence for green fluorescent protein (GFP) was fused in frame to the amino-terminal (N-terminal) coding sequence of AtNACK1/HIK cDNA (Fig. 1b). This fusion gene was driven by the estrogen-inducible promoter XVE (Zuo et al. 2000) and introduced into cells of the tobacco cultured-cell line BY-2. We cultured the transformed BY-2 cells in liquid medium containing 0.1 µM 17-β-estradiol for 16 h to induce expression of GFP-AtNACK1. We observed subcellular localization of GFP-AtNACK1 in the transformed cells (Fig. 2), where it appeared in the central region corresponding to the expanding cell plate. The GFP signals disappeared as the formation of cell plates ended, suggesting that AtNACK1 disappears upon the maturation of cell plates. These results are consistent with the pattern of a sharp accumulation of tobacco NACK1 protein seen at the M phase of the cell cycle (Nishihama et al. 2002).
Subcellular localization patterns of GFP-AtNACK1 during the formation of cell plates in BY-2 cells. BY-2 cells transformed with estradiol-inducible GFP-AtNACK1 were incubated for 16 h in the presence of 0.1 µM 17-β-estradiol. Living cells expressing GFP-AtNACK1 were observed by confocal fluorescence microscopy to detect fluorescence of sGFP (upper) and recorded as time-lapse micrographs at the indicated times during expansion of the cell plate. Normarski (DIC) and merged images (merged) are shown in the center and lower panels, respectively. Bar 10 µm
The amino-terminal motor and the carboxyl-terminal region of the stalk of AtNACK1/HIK are required for its localization to the site of cell plate formation
To examine which sequences of AtNACK1/HIK are involved in the localization to the site of cell plate formation, we made a series of deletion constructs of AtNACK1 cDNA (Fig. 1b), which contained the DNA sequence corresponding to the full length cDNA (full), the motor domain (MD), the stalk (ST) sequence, the MD-A sequence covering the MD and A regions in ST, the MD-AB sequence covering MD, the A and B regions in ST, the MD-AC sequence covering MD, the A and C regions in ST, and the MD-BC sequence covering the MD and BC regions in ST. Note that the B region contained the predicted binding sites of ANP1 MAPKKKs and the putative CDK phosphorylation sites (Ishikawa et al. 2002; Takahashi et al. 2010). These DNA constructs were fused to the C-terminus sequence of the GFP gene, whose expression was under the control of the constitutive active Cauliflower mosaic virus (CaMV) 35S promoter, and the chimeric DNA constructs were introduced into cells of the cultured tobacco cell line BY-2.
We observed GFP signals during cytokinesis in transformed BY-2 cells (Fig. 3). In cells expressing GFP-AtNACK1:MD, no GFP signals were observed in either the phragmoplast region or the site of cell plate formation (Fig. 3b). Although we also transformed BY-2 cells with the GFP-AtNACK1:ST and obtained some calli, these calli were unable to grow further (data not shown), which might be due to the mislocalization of the motor-less NACK1:ST, because this observation is consistent with the result that the overexpression of the stalk region of tobacco NACK1 prevents NPK1 MAPKKK from locating to the phragmoplast equator and has a dominant-negative effect (Nishihama et al. 2002). Taken together, the motor domain seems to play a critical role in the AtNACK1 localization to the site of cell plate formation, but only the motor domain is not sufficient for its proper localization.
Subcellular localization of GFP-AtNACK1 and its derivatives during formation of cell plates in BY-2 cells. Expression of indicated constructs of AtNACK1 was controlled under the 35S promoter. Transformed BY-2 cells expressing GFP-AtNACK1 (a), GFP-AtNACK1:MD (b), GFP-AtNACK1:MD-A (c), GFP-AtNACK1:MD-AB (d), GFP-AtNACK1:MD-AC (e), and GFP-AtNACK1:MD-BC (f) were observed by fluorescence microscopy to detect fluorescence of G3GFP (center) during cell plate formation. Nomarski images (DIC) and merged images (merged) are shown in the left and right panels, respectively. Arrowheads indicate the site of expanding cell plates. Bar 20 µm
GFP signals were not detected at the site of cell plate formation in the transformed cells with GFP-AtNACK1:MD-A, GFP-AtNACK1:MD-AB, and GFP-AtNACK1:MD-AC (Fig. 3c–e). Obvious GFP signals were observed, however, at the cell plate formation site in the transformed BY-2 cells expressing GFP-AtNACK1:MD-BC (Fig. 3f). These results indicate that the BC region in ST, in addition to the motor domain of AtNACK1/HIK, is sufficient for the localization of AtNACK1 to the site of cell plate formation, but that only the AB region and the C region when separated from the B region are not sufficient for the appropriate localization. These results also indicate that the A region is not required for its localization to the site of cell plate formation.
AtNACK1-GFP fusion does not localize to the site of cell plate formation in the living cells of Arabidopsis roots and causes dominant-negative effects on cytokinesis in Arabidopsis plants
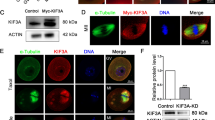
We fused the C-terminal coding sequence of the entire AtNACK1/HIK genomic sequence, which covered the 5′-upstream promoter and coding regions, to the N-terminus of the GFP sequence. The genomic sequence we used has the ability to complement the phenotypes of atnack1 mutants (pAtNACK1::genome AtANCK1; Sasabe et al. 2011a). We used this construct to transform the wild-type Arabidopsis plants (Col-0). We observed the GFP signals by means of confocal laser scanning microscopy. By analyzing time-lapse images of living cells in the transgenic plants (Fig. 4a), we observed that the signals derived from pAtNACK1::genome AtNACK1-GFP did not localize at the site of cell plate formation, as visualized by staining with FM4–64, but were detected within wide areas corresponding to the whole phragmoplast in transformed root cells. The cell plate stained by FM4–64 in the transgenic Arabidopsis expressing AtNACK1-GFP appeared to expand more slowly than that in non-transgenic Arabidopsis (Fendrych et al. 2010; Kosetsu et al. 2010). These results indicate that the C-terminal GFP fusion of AtNACK1 appears to interfere with its localization to the site of cell plate formation, suggesting that the C-terminal region of AtNACK1/HIK plays an important role in its localization to the site of cell plate formation as well as in the cell plate formation itself.
Subcellular localization of AtNACK1-GFP during the formation of cell plates in Arabidopsis plants and BY-2 cells, and effects of its expression on the plant phenotypes. a Time-lapse micrographs of AtNACK1-GFP in the living root-tip cell of an Arabidopsis plant that was transformed with genomic AtNACK1-fused GFP. Genomic AtNACK1 constructs were controlled under the original promoter. Confocal images were recorded at indicated times after the detection of cell plate formation. Fluorescence images show the signals due to FM4–64 (left, magenta) and GFP (center, green). The merged images (merged) are shown in the right panels. Arrowheads indicate the site of expanding cell plates. Bar 10 µm. b Comparison of gross morphology of one 14-day-old wild type Col-0 plant and three T2 plants transformed with genomic AtNACK1-fused GFP. T2 plant line 1 (#1) showed the most severe dwarf phenotype. Bar 10 mm. c Cytokinetic defects in epidermal cells of leaves in transgenic plants. Fourteen-day-old Col-0 plants and T2 plants, line 1, were cleared and the epidermal cells in their leaves were observed. Arrows indicate the incomplete cell walls. Bars 10 µm. d Comparison of gross morphology of 46-day-old wild type Col-0 plant and T3 plant transformed with genomic AtNACK1-fused GFP. These plants were grown on soil under the long-day condition. T3 plant line 8 (#8) that homozygously expressed AtNACK1-GFP showed dwarf phenotypes at an efficiency of 8 %. Bars 10 mm. e–g Comparison of the patterns of subcellular localization of AtNACK1-GFP and GFP-AtNACK1 in BY-2 cells. Expression of GFP-AtNACK1 (e) and AtNACK1-GFP (f, g) were induced by 0.1 µM estradiol and fluorescence due to GFP was observed by fluorescence microscopy during cell plate formation. Fluorescence images (GFP), Nomarski images (DIC) and merged images (merged) in the cell exhibiting the representative patterns are shown. Images observed in transformed lines 16 (f, #16) and 43 (g, #43) are shown. Arrowheads indicate the sites of expanding cell plates. Bar 20 µm
We observed the phenotypes of Arabidopsis plants transformed with pAtNACK1::genome AtNACK1-GFP (Fig. 4b–d). In T2 plants, 6 out of 7 transgenic lines displayed a dwarf phenotype (Fig. 4b; 6–72 %), although the extent of the phenotype depended on each transgenic line. Epidermal cells in the #1 transgenic line, which displayed the highest proportion (72 %) of the most severe dwarf phenotype, exhibited cytokinesis defects with incomplete cell walls (Fig. 4c). Although the #8 transgenic plant line did not show dwarfism in T2 plants, the T3 plants homozygously expressing AtNACK1-GFP showed 8 % of dwarf phenotypes (Fig. 4d, nested image; 4 of 50 seedlings). The dwarf phenotypes might be due to a growth defect of the transgenic plants owing to the mislocalization of AtNACK1-GFP as described above.
AtNACK1-GFP localizes to the site of cell plate formation and the wide area outside of the site in BY-2 cells
We also examined subcellular localization of AtNACK1-GFP in BY-2 cells and compared the pattern of its localization with that of the pattern of localization of AtNACK1-GFP in Arabidopsis. We made the XVE::AtNACK1-GFP construct, in which the GFP sequence was fused to the C-terminal coding sequence of AtNACK1 cDNA, and introduced into BY-2 cells. We cultured the transformed BY-2 cells in liquid medium containing 0.1 µM estradiol for 16 h to induce expression of AtNACK1-GFP. As compared with the GFP signal derived from XVE::GFP-AtNACK1 (Fig. 4e), those derived from XVE::AtNACK1-GFP were detected in the wide regions around the site of cell plate formation (Fig. 4f, g), which might have corresponded to the phragmoplast area. These results indicate that the C-terminal GFP fusion of AtNACK1 appears to disturb its localization to the site of cell plate formation in BY-2 cells as well as in Arabidopsis plants. We have also made the AtNACK1-GFP construct with a constitutive active CaMV 35S promoter and have attempted to analyze the subcellular localization of AtNACK1-GFP in tobacco BY-2 cells. Although we obtained transformed BY-2 cells, we failed to detect GFP signals.
Discussion
AtNACK1/HIK is localized to the cell plate formation site in the phragmoplast of tobacco BY-2 cells and degraded after cell plate formation
The results of the present study with the GFP-AtNACK1/HIK construct have shown that the GFP-AtNACK1 protein is localized to the site of cell plate formation during cytokinesis in tobacco BY-2 cells (Fig. 2). The GFP signals disappeared when the signals reached the parental cell walls (Fig. 2), suggesting that the fusion protein degrades after cell plate formation, which is consistent with the previous results obtained by western blot analysis: tobacco NACK1 protein is accumulated specifically during M phase and disappears at the end of the M phase in synchronized BY-2 cells (Nishihama et al. 2002). The M-phase-specific accumulation partially depends on the specific transcription at the M phase by the Myb transcription factors (Ito et al. 2001; Araki et al. 2004). The specific accumulation should also be due to the disappearance of NACK1 protein around the end of M phase, which is inhibited by the persistent CDK activity (Sasabe et al. 2011a) and the treatment with MG-132, a proteasome inhibitor (our unpublished data), suggesting that NACK1 is degraded by the action of the ubiquitin–proteasome pathway. AtNACK1 of Arabidopsis might be degraded by a similar mechanism at the end of cytokinesis in tobacco BY-2 cells (Fig. 2). The mechanism involved in this phase-specific degradation of NACK proteins should be further investigated experimentally.
We also transformed Arabidopsis plants with the same GFP-AtNACK1 construct, and obtained sufficient numbers of transgenic plant lines. We, however, failed to find plants that were GFP-positive after induction with estrogen.
The BC region in the stalk of AtNACK1/HIK is active in its binding to phragmoplast microtubules and localizing to the site of cell plate formation
The sequences covering the motor domain and the BC region of the stalk of AtNACK1 (Fig. 1) are sufficient for its localization to the site of cell plate formation in the phragmoplast, but the A region is not required (Fig. 3). Characteristic features of the BC region in AtNACK1 are the presence of three predicted coiled-coil structures (4th, 5th, and 6th positions of coiled-coils in Fig. 1a), one of which overlaps with the site of interaction with ANP1 MAPKKK (corresponding to NPK1 in tobacco), a component of the PQR MAPK pathway that positively controls the formation of cell plates (Ishikawa et al. 2002; Kosetsu et al. 2010; Nishihama et al. 2002; Sasabe et al. 2011a; Takahashi et al. 2010). Considering the general concepts of configurations and dynamics of kinesin motors (Seeger and Rice 2010; Sablin 2000; Verhey and Hammond 2009), these coiled-coil segments together with the motor domain of AtNACK1 possibly play at least two critical roles in their specific localization. First, these coiled-coil structures might be involved in oligomerization of AtNACK1 kinesin, which might be required for their binding to microtubules and motility along the microtubules to move to the site for cell plate formation. The motility property is generated by coordinate stepping of at least two motor domains in the kinesin oligomers.
Second, the coiled-coil structures of the BC region might provide binding sites for cargos to be transported along the microtubules, and the binding of the AtNACK1 kinesin motor to microtubules and its motility activity on the microtubule might be stimulated by the interaction with the PQR MAPK pathway similarly as by interactions between motors and cargos of other known kinesins. Kinesins, in general, bind to proper cargos at their non-motor regions, such as stalks and C-terminal tail regions, and transport the cargos along microtubule filaments to genetically determined destinations in specific subcellular compartments where they exert their precise functions (Verhey and Hammond 2009; Verhey et al. 2011). In the absence of cargos, kinesins are inactive, in terms of binding to microtubules and motility activities, due to their conformational repressive states, which have been recently defined as autoinhibition mechanisms (Hammond et al. 2010; Kaan et al. 2011; Verhey and Hammond 2009; Verhey et al. 2011). Since the complex of components in the PQR MAPK pathway binds to the B region of the AtNACK1 stalk (Ishikawa et al. 2002; Takahashi et al. 2010), the protein complex might release the autoinhibition of AtNACK1 to bind microtubules and activate its motility function. A well-characterized example is the cargo protein of the mammalian Kinesin-1 motor, which is the JIP scaffold protein that assembles the JNK MAPK pathway consisting of MAPKKK, MAPKK, and JNK MAPK and activates the Kinesin-1 motor (Blasius et al. 2007; Verhey et al. 2001). Activation of the JNK pathway seems to negatively regulate the interaction of kinesin motors with their cargos (Horiuchi et al. 2007), suggesting the presence of crosstalk between the MAPK signaling and trafficking pathways. It must be worth testing whether similar crosstalk between PQR MAPK and the vesicle trafficking systems modulated by AtNACK1 kinesin might be involved in the formation of cell plates.
The C-terminus of AtNACK1/HIK is involved in the processive motility of AtNACK1/HIK on microtubules
It has been reported that Myc-tagged AtNACK1/HINKEL proteins expressed under the original promoter are localized at the midzone of dividing cells in Arabidopsis plants (Krupnova et al. 2013). AtNACK1-GFP proteins that were synthesized in the Arabidopsis plant from the pAtNACK1::genome AtNACK1-GFP construct and in BY-2 cells from the XVE::AtNACK1-GFP construct were not clearly localized to the site of cell plate formation, however, but rather distributed over wide areas, which appear to correspond to phragmoplast microtubules, around the newly formed midzone (Fig. 4a, f, g). Considering the potential microtubule-binding activity and the motor activity of NACK proteins (Nishihama et al. 2002), the observed diffusive localization of AtNACK1-GFP might be due to inhibitory effects of GFP, which had been joined to the C-terminus of AtNACK1, on its motility activity; however, AtNACK1-GFP might still maintain the binding activity to microtubules because the GFP signals appear to localize on the phragmoplast area (Fig. 4a, f, g). Within the population of these transgenic plants expressing pAtNACK1::genome AtNACK1-GFP, we observed plants showing the dwarf phenotype and cytokinetic defects (Fig. 4b–d), suggesting that the AtNACK1-GFP protein has a dominant-inhibitory effect on wild-type AtNACK1 in the transgenic plants. The Arabidopsis mutation of RUNKEL exhibits severe cytokinetic defects and results in seedling lethal (Krupnova et al. 2009). Recently, it has been reported that the transgenic plants expressing truncated RUNKEL proteins cause the mislocalization of Myc-AtNACK1/HIK, such that it is dispersed around the cell plate and between the daughter nuclei from early to late stages of cytokinesis in these plants (Krupnova et al. 2013). Again, GFP fusion to the C-terminus of AtNACK1 might affect the interaction between AtNACK1 and RUNKEL, or other unknown cargos such as vesicles to be required for the cell plate formation, which might cause the mislocalization of AtNACK1-GFP during cell plate formation. Functions of NACK kinesins and their regulatory mechanisms in the formation of cell plates should be further investigated.
References
An G (1985) High efficiency transformation of cultured tobacco cells. Plant Physiol 79:568–570
Araki S, Ito M, Soyano T, Nishihama R, Machida Y (2004) Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem 279:32979–32988
Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ (2007) Two binding partners cooperate to activate the molecular motor kinesin-1. J Cell Biol 176:11–17
Chow CM, Neto H, Foucart C, Moore I (2008) Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20:101–123
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Fendrych M, Synek L, Pecenková T, Toupalová H, Cole R, Drdová E, Nebesárová J, Sedinová M, Hála M, Fowler JE, Zársky V (2010) The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22:3053–3065
Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ (2010) Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol 189:1013–1025
Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM (2007) Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol 17:1313–1317
Ishikawa M, Soyano T, Nishihama R, Machida Y (2002) The NPK1 mitogen-activated protein kinase kinase kinase contains a functional nuclear localization signal at the binding site for the NACK1 kinesin-like protein. Plant J 32:789–798
Ishikawa T, Machida C, Yoshioka Y, Kitano H, Machida Y (2003) The GLOBULAR ARREST1 gene, which is involved in the biosynthesis of folates, is essential for embryogenesis in Arabidopsis thaliana. Plant J 33:235–244
Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13:1891–1905
Jürgens G (2005) Plant cytokinesis: fission by fusion. Trends Cell Biol 15:277–283
Kaan HY, Hackney DD, Kozielski F (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333:883–885
Kosetsu K, Matsunaga S, Nakagami H, Colcombet J, Sasabe M, Soyano T, Takahashi Y, Hirt H, Machida Y (2010) The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22:3778–3790
Krupnova T, Sasabe M, Ghebreghiorghis L, Gruber CW, Hamada T, Dehmel V, Strompen G, Stierhof YD, Lukowitz W, Kemmerling B, Machida Y, Hashimoto T, Mayer U, Jürgens G (2009) Microtubule-associated kinase-like protein RUNKEL needed for cell plate expansion in Arabidopsis cytokinesis. Curr Biol 19:518–523
Krupnova T, Stierhof YD, Hiller U, Strompen G, Müller S (2013) The microtubule-associated kinase-like protein RUNKEL functions in somatic and syncytial cytokinesis. Plant J 74:781–791
Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139:1485–1493
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
McMichael CM, Bednarek SY (2013) Cytoskeletal and membrane dynamics during higher plant cytokinesis. New Phytol 197:1039–1057
Miki T, Naito H, Nishina M, Goshima G (2014) Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc Natl Acad Sci USA 111:E1053–E1061
Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14:412–417
Müller S, Wright AJ, Smith LG (2009) Division plane control in plants: new players in the band. Trends Cell Biol 19:180–188
Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4:1967
Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71:2095–2100
Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, Obuse C, Kubo M, Hiwatashi Y, Goshima G (2012) An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 24:1478–1493
Nishihama R, Machida Y (2001) Expansion of the phragmoplast during plant cytokinesis: a MAPK pathway may MAP it out. Curr Opin Plant Biol 4:507–512
Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y (2001) The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev 15:352–363
Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, Yamazaki Y, Machida Y (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109:87–99
Normand G, King RW (2010) Understanding cytokinesis failure. Adv Exp Med Biol 676:27–55
Otegui MS, Verbrugghe KJ, Skop AR (2005) Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol 15:404–413
Peterman TK, Ohol YM, McReynolds LJ, Luna EJ (2004) Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol 136:3080–3094
Sablin EP (2000) Kinesins and microtubules: their structures and motor mechanisms. Curr Opin Cell Biol 12:35–41
Sasabe M, Soyano T, Takahashi Y, Sonobe S, Igarashi H, Itoh TJ, Hidaka M, Machida Y (2006) Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev 20:1004–1014
Sasabe M, Boudolf V, De Veylder L, Inzé D, Genschik P, Machida Y (2011a) Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc Natl Acad Sci USA 108:17844–17849
Sasabe M, Kosetsu K, Hidaka M, Murase A, Machida Y (2011b) Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal Behav 6:743–747
Sazuka T, Aichi I, Kawai T, Matsuo N, Kitano H, Matsuoka M (2005) The rice mutant dwarf bamboo shoot 1: a leaky mutant of the NACK-type kinesin-like gene can initiate organ primordia but not organ development. Plant Cell Physiol 46:1934–1943
Seeger MA, Rice SE (2010) Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem 285:8155–8162
Smertenko AP, Piette B, Hussey PJ (2011) The origin of phragmoplast asymmetry. Curr Biol 21:1924–1930
Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 17:1055–1067
Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U (2002) The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12:153–158
Takahashi Y, Soyano T, Kosetsu K, Sasabe M, Machida Y (2010) HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol 51:1766–1776
Tanaka H, Ishikawa M, Kitamura S, Takahashi Y, Soyano T, Machida C, Machida Y (2004) The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9:1199–1211
Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, Hussey PJ (2002) MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol 4:711–714
Van Damme D, Geelen D (2008) Demarcation of the cortical division zone in dividing plant cells. Cell Biol Int 32:178–187
Verhey KJ, Hammond JW (2009) Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol 10:765–777
Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B (2001) Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol 152:959–970
Verhey KJ, Kaul N, Soppina V (2011) Kinesin assembly and movement in cells. Annu Rev Biophys 40:267–288
Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jürgens G, Mayer U (2000) The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr Biol 10:1371–1374
Woollard AA, Moore I (2008) The functions of Rab GTPases in plant membrane traffic. Curr Opin Plant Biol 11:610–619
Yasuhara H, Sonobe S, Shibaoka H (1993) Effects of taxol on the development of the cell plate and of the phragmoplast in tobacco BY-2 cells. Plant Cell Physiol 34:21–29
Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24:265–273
Acknowledgments
This work was supported, in part, by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 25114504) from JSPS to M.S., by a Grant-in-Aid for Scientific Research on Priority Areas (No. 19060003) to Y.M., by a Grant-in-Aid for Young Scientists (B; No. 21770040 and No. 26840086) to M.S. and by a Grant-in-Aid for Scientific Research (B; No. 23370021) to Y.M. and M.I. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported, in part, by a Hirosaki University Grant for Exploratory Research by Young Scientists to M.S., by Grant for Basic Science Research Projects from The Sumitomo Foundation to M.S. and by a Hirosaki University Institutional Research Grant for Young Scientists. Microscopy was conducted using LSM780 at the Institute of Transformative Bio-Molecules at Nagoya University and supported by the Japan Advanced Plant Science Research Network.
Author information
Authors and Affiliations
Corresponding authors
Additional information
M. Sasabe and N. Ishibashi are contributed equally to the preparation of this manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sasabe, M., Ishibashi, N., Haruta, T. et al. The carboxyl-terminal tail of the stalk of Arabidopsis NACK1/HINKEL kinesin is required for its localization to the cell plate formation site. J Plant Res 128, 327–336 (2015). https://doi.org/10.1007/s10265-014-0687-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-014-0687-2