Abstract
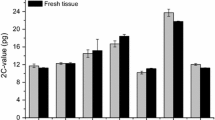
The genome size was surveyed in 13 Notolathyrus species endemic to South America by flow cytometry and analyzed in an evolutionary and biogeographic context. A DNA content variation of 1.7-fold was registered, and four groups of species with different DNA content were determined. Although, the 2C values were correlated with the total chromosome length and intrachromosomal asymmetry index (A1), the karyotype formula remained almost constant. The conservation of the karyotype formula is in agreement with proportional changes of DNA in the chromosome arms. Species with annual life cycle and shorter generation time had the lowest DNA content and the data suggest that changes in DNA content involved reductions of genome size in the perennial to annual transitions. The variation of 2C values was correlated with precipitation of the coldest quarter and, to some extent, with altitude. Additional correlations with other variables were observed when the species were analyzed separately according to the biogeographic regions. In general, the species with higher DNA content were found in more stable environments. The bulk of evidence suggests that changes on genome size would have been one of the most important mechanisms that drove or accompanied the diversification of Notolathyrus species.




Similar content being viewed by others
References
Albach DC, Grilhuber J (2004) Genome size variation and evolution in Veronica. Ann Bot 94:897–911
Ali HBM, Meister A, Schubert I (2000) DNA content, rDNA loci, and DAPI bands reflect the phylogenetic distance between Lathyrus species. Genome 43:1027–1032
Ayaz E, Ertekin SA (2008) Karyotype analysis of two species of genus Lathyrus from South-eastern Anatolia, Turkey. Int J Agric Biol 5:569–572
Bancheva S, Greilhuber J (2006) Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Syst Evol 257:95–117
Baranyi M, Greilhuber J (1999) Genome size in Allium: in quest of reproducible data. Ann Bot 83:687–695
Battistin A, Fernández A (1994) Karyotypes of four species of South America natives and one cultivated species of Lathyrus L. Caryologia 47:325–330
Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proc R Soc Lond B 181:109–135
Bennett MD (1976) DNA amount, latitude, and crop plant distribution. Environ Exp Bot 16:93–108
Bennett MD (1982a) Nucleotypic basis of the spatial ordering of the chromosomes in eukaryotes and implications of the order for genome evolution and phenotypic variation. In: Dover GA, Flavell RB (eds) Genome evolution. Academic Press, London and New York
Bennett MD (1982b) The spatial distribution of chromosomes. In: Brandham PE, Bennett MD (eds) Kew chromosome conference II. The Royal Botanic Gardens, Kew, pp 71–79
Bennett MD (1987) Variation in genomic form in plants and its ecological implications. New Phytol 106:177–200
Bennett MD (1995) The development and use of genomic in situ hybridization (GISH) as a new tool in plant biosystematics. In: Brandham PE, Bennett MD (eds) Kew chromosome conference IV. The Royal Botanic Gardens, Kew, pp 167–183
Bennett MD, Leitch IJ (2005) Genome size evolution in plants. In: Gregory TR (ed) The evolution of the genome 2:89–162. Elsevier Academics, New York
Bennett MD, Smith JB (1991) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B 334:309–345
Bennett MD, Johnston S, Hodnett GL, Price HJ (2000) Allium cepa L. cultivars from four continents compared by flow cytometry show nuclear DNA constancy. Ann Bot 85:351–357
Bennetzen JL (2002) Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115:29–36
Bennetzen JL, Kellogg EA (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell 9:1509–1514
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Bottini MCJ, Greizerstein EJ, Aulicino MB, Poggio L (2000) Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW patagonian species of Berberis L. (Berberidaceae). Ann Bot 86:565–573
Brandham PE, Doherty MJ (1998) Size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot 82:67–73
Burkart A (1935) Revision of the Lathyrus species from Argentina Republic (in Spanish). R Fac Agron Univ Nac La Plata 8:41–128
Burkart A (1942) New contributions to the systematics of the South American species of Lathyrus (in Spanish). Darwiniana 6:9–29
Caceres ME, De Pace C, Scarascia Mugnozza GT, Kotsonis P, Ceccarelli M, Cionini PG (1998) Genome size variation within Dasypyrum villosum: correlation with chromosomal traits, environmental factors and plant phenotypic characteristics and behavior in reproduction. Theor Appl Genet 96:559–567
Chalup L, Grabiele M, Solís Neffa V, Seijo G (2012) Structural karyotypic variability and polyploidy in natural populations of the South American Lathyrus nervosus Lam. (Fabaceae). Plant Syst Evol 298:761–773
Choi WY (1971) Variation in nuclear DNA content in the genus Vicia. Genetics 68:195–211
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Di Rienzo JA, Casanoves F, Balzarini MG, González L, Tablada M, Robledo CW (2013) InfoStat version 2013. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar
Doležel J, Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot 95:99–110
Doležel J, Greilhuber RJ, Suda J (2007) Estimation of nuclear DNA content in plant using flow cytometry. Nat Protoc 2:2233–2244
Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131:900–910
Ghasem K, Danesh-Gilevaei M, Aghaalikhani M (2011) Karyotypic and nuclear DNA variations in Lathyrus sativus (Fabaceae). Caryologia 64:42–54
Gregory TR (2001) Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev 76:65–101
Gregory TR (2004) Insertion-deletion biases and the evolution of genome size. Gene 324:15–34
Gregory TR (2005a) Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6:699–708
Gregory TR (2005b) The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot 95:133–143
Greilhuber J (1998) Intraspecific variation in genome size: a critical reassessment. Ann Bot 82(Suppl. A):27–35
Greilhuber J (2005) Intraspecific variation in genome size in angiosperms: identifying its existence. Ann Bot 95:91–98
Grime JP, Mowforth MA (1982) Variation in genome size an ecological interpretation. Nature 299:151–153
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Jakob SS, Meister A, Blattner FR (2004) The considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Mol Biol Evol 21:860–869
Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ (2005) Evolution of genome size in Brassicaceae. Ann Bot 95:229–235
Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607
Kenicer GJ, Kajita T, Pennington RT, Murata J (2005) Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. Am J Bot 92:1199–1209
Klamt A, Schifino-Wittmann MT (2000) Karyotype morphology and evolution in some Lathyrus (Fabaceae) species of southern Brazil. Genet Mol Biol 23:463–467
Knight CA, Ackerly DD (2002) Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecol Lett 5:66–76
Knight CA, Molinari NA, Petrov DA (2005) The large genome constraint hypothesis: evolution, ecology and phenotype. Ann Bot 95:177–190
Krapovickas A, Fuchs AM (1957) Cytological notes in legumes (in Spanish). II. R Investig Agric 11:215–218
Kron P, Suda J, Husband BC (2007) Applications of flow cytometry to evolutionary and population biology. Annu Rev Ecol Evol Syst 38:847–876
Kupicha FK (1983) The infrageneric structure of Lathyrus. Notes R Bot Gard Edinb 41:209–244
Levin DA, Funderburg SW (1979) Genome size in angiosperms: temperate versus tropical species. Am Nat 114:784–795
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Mac Gillivray CW, Grime JP (1995) Genome size predicts frost resistance in British herbaceous plants: implications for rates of vegetation response to global warming. Funct Ecol 9:320–325
Moscone EA, Baranyi M, Ebert L, Greilhuber L, Ehrendorfer F, Hunziker AT (2003) Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Ann Bot 92:21–29
Nandini AV, Murray BG (1997) Intra and interspecific variation in genome size in Lathyrus (Leguminosae). Bot J Linn Soc 125:359–366
Naranjo CA, Ferrari MR, Palermo AM, Poggio L (1998) Karyotype DNA content and meiotic behaviour in five South American species of Vicia (Fabaceae). Ann Bot 82:757–764
Narayan RKJ (1982) Discontinuous DNA variation in the evolution of plant species: the genus Lathyrus. Evolution 36:877–891
Narayan RKJ (1998) The role of genomic constraints upon evolutionary changes in genome size and chromosome organization. Ann Bot 82:57–66
Narayan RKJ, Durrant A (1983) DNA distribution in chromosomes of Lathyrus species. Genetica 61:47–53
Narayan RKJ, McIntre FK (1989) Chromosomal DNA variation, genomic constraints and recombination in Lathyrus. Genetica 79:45–52
Ohri D, Khoshoo TN (1986) Genome size in gymnosperms. Plant Syst Evol 153:119–132
Petrov DA (2002) Mutational equilibrium model of genome size evolution. Theor Popul Biol 61:531–543
Petrov DA, Hartl DL (1997) Trash DNA is what gets thrown away: high rate of DNA loss in Drosophila. Gene 205:279–289
Poggio L, Naranjo CA (1990) DNA content and evolution in higher plants (in Spanish). Acad Nac Ex Fis Nat Buenos Aires Monografía 5:27–37
Poggio L, Burghardt AD, Hunziker JH (1989) Nuclear DNA variation in diploid and polyploid taxa of Larrea (Zygophyllaceae). J Hered 63:321–328
Poggio L, Rosato M, Chiavarino AM, Naranjo C (1998) Genome size and environmental correlations in Maize (Zea mays ssp. mays, Poaceae). Ann Bot 82:107–115
Poggio L, Espert SM, Fortunato RH (2008) Evolutive cytogenetics in American legumes (in Spanish). Rodriguesia 59:423–433
Price HJ (1988) Nuclear DNA content variation within angiosperm species. Evol Trends Plants 2:53–60
Price HJ, Bachmann K (1975) DNA content and evolution in the Microseridinae. Am J Bot 62:262–267
Price HJ, Johnston JS (1996) Influence of light on DNA content of Helianthus annuus Linnaeus. Proc Natl Acad Sci USA 93:11264–11267
Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, Johnston JS (2005) Genome evolution in the genus Sorghum (Poaceae). Ann Bot 95:219–227
Rayburn AL, Auger JA (1990) Genome size variation in Zea mays spp mays adapted to different altitudes. Theor Appl Genet 79:470–474
Rees H, Hazarika MH (1969) Chromosome evolution in Lathyrus. In: Darlington CD, Lewis KR (eds) Chromosomes today, vol 2. New York, pp 158–165
Rees H, Narayan GH (1988) Chromosome constrains: chiasma frequency and genome size. In: Brandham PE (ed) Kew chromosome conference III. The Royal Botanic Gardens, Kew, pp 231–240
Schmuths H, Meister A, Horres R, Bachmann K (2004) Genome size variation among accessions of Arabidopsis thaliana. Ann Bot 93:317–321
Schweizer D, Loidl J (1987) A model for heterochromatin dispersion and the evolution of C-band patterns. In: Sthal A, Luciani JM, Vagner-Capodano AM (eds) Chromosomes today, vol 9. Marseille, pp 61–74
Seal AG (1983) DNA variation in Festuca. Heredity 50:225–236
Seal AG, Rees H (1982) The distribution of quantitative DNA changes associated with the evolution of diploid Festuceae. Heredity 49:179–190
Seijo JG (2002) Cytogenetic studies in South American species of the genus Lathyrus, section Notolathyrus (Leguminosae) (in Spanish). PhD thesis, National University of Córdoba, Argentina
Seijo JG, Fernández A (2003) Karyotype analysis and chromosome evolution in South American species of Lathyrus (Leguminosae). Am J Bot 90:980–987
Seijo JG, Solís Neffa VG (2006) Cytogenetic studies in the rare South American Lathyrus hasslerianus Burk. Cytologia 71:11–19
Sims LE, Price HJ (1985) Nuclear DNA content variation in Helianthus annuus (Asteraceae). Am J Bot 72:1213–1219
Soltis DE, Soltis PS, Tate JA (2003) Advances in the study of polyploidy since plant speciation. New Phytol 161:173–191
Stebbins GL (1957) Self-fertilization and population variability in the higher plants. Am Nat 91:337–354
Temsch EM, Greilhuber J (2000) Genome size variation in Arachis hypogaea and A. monticola re-evaluated. Genome 43:449–451
Temsch EM, Greilhuber J (2001) Genome size in Arachis duranensis: a critical study. Genome 44:826–830
Thiers B (2013) Index herbariorum: a global directory of public herbaria and associated staff. Botanical Garden’s Virtual Herbarium, New York. http://sweetgum.nybg.org/ih
Wakamiya I, Newton R, Johnston JS, Price HJ (1993) Genome size and environmental factors in the genus Pinus. Am J Bot 80:1235–1241
Wendel JF, Cronn RC, Johnston JS, Price HJ (2002) Feast and famine in plant genomes. Genetica 115:37–47
Wicker T, Guyot R, Yahiaoui N, Keller B (2003) CACTA transposons in Triticeae. A diverse family of high-copy repetitive elements. Plant Physiol 132:52–63
Yamamoto K, Fujiwara T, Blumenreich ID (1984) Karyotypes and morphological characteristics of some species in the genus Lathyrus L. J Breed 34:273–284
Acknowledgments
This research was partially supported by grants of Agencia Nacional de Promoción Científica, Tecnológica y de Innovación (ANPCyT-FONCyT-UNNE PICTO 2007-90). Collection trips were partially supported by the Myndel Botanica Foundation. L. Chalup is a Doctoral Fellow of the National Research Council of Argentina (CONICET), J.G. Seijo and V.G. Solís Neffa are members of the Carrera del Investigador Científico of CONICET.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chalup, L., Grabiele, M., Neffa, V.S. et al. DNA content in South American endemic species of Lathyrus . J Plant Res 127, 469–480 (2014). https://doi.org/10.1007/s10265-014-0637-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-014-0637-z