Abstract
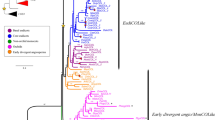
A nuclear gene, FLOWERING LOCUS T (FT) homolog, was cloned from Phyllostachys meyeri as PmFT. Its putative copy number was estimated as four by Southern blot analysis, and the two copies were completely sequenced. Twenty-seven FT homolog sequences of bambusoid and early diverging grasses comprised 172-bp exons, and 357- to 785-bp introns exhibited 0–58.9% pairwise divergence with six modal levels. Parsimony analyses of the FT homologs rooted at Pharus virescens produced six equally parsimonious trees. In the strict consensus tree, five clades were resolved; they were affected by divergence of the intron region rather than exon region. The basal clade was Puelioideae, followed by Olyreae clade including Oryza sativa. Streptogyneae clade combined the Olyreae clade with terminal sister clades of the Bambuseae, i.e., pantropical bamboos and East Asiatic temperate bamboos. The global topology suggested that FT homologs are significant for resolving the tribe level. However, the phylogeny of FT homologs does not resolve monophyly in Bambusoideae because of intercalary positioning by Streptogyneae clade. We discussed the role of FT homologs in controlling the inflorescence architecture and position of Streptogyneae in the bamboo phylogeny.




Similar content being viewed by others
References
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379:791–797
Chardon F, Damerval C (2005) Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol 61:579–590
Clark LG, Zhang W, Wendel JF (1995) A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Syst Bot 20:436–460
Clark LG, Kobayashi M, Mathews S, Spangler RE, Kellogg EA (2000) The Puelioideae, a new subfamily of Poaceae. Syst Bot 25:181–187
Clayton WD, Renvoize SA (1986) Genera graminum: grasses of the world. Her Majesty’s Stationery Office, London
GPWG (2000) A phylogeny of the grass family (Poaceae), as inferred from eight character sets. In: Jacob SWL, Everett J (eds) Grasses: systematic and evolution. CSIRO, Melbourne, pp 3–7
GPWG (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88:373–457
Guo ZH, Li DZ (2004) Phylogenetics of the Thamnocalamus group and its allies (Gramineae: Bambusoideae): inference from the sequences of GBSSI gene and ITS spacer. Mol Phylogenet Evol 30:1–12
Harper JL (1977) Population biology of plants. Academic Press, London, p 892
Hasebe M, Iwatsuki K (1990) Adiantum capillus-veneris chloroplast DNA clone bank as useful heterologous probes in the systematics of the leptosporangiate ferns. Am Fern J 80:20–25
Hisamoto Y, Kobayashi M (2007) Comparison of nucleotide sequences of fragments from rice FLOWERING LOCUS T (RFT1) homologs in Phyllostachys (Bambusoideae, Poaceae) with particular reference to flowering behaviour. Kew Bull 62:463–473
Hisamoto Y, Kashiwagi H, Kobayashi M (2005) Monocarpic mass flowering and flower morphology of Phyllostachys meyeri McClure (Poaceae: Bambusoideae) cultured in the Fuji Bamboo Garden, Japan. J Jpn Bot 80:63–71
Izawa K, Kobayashi M (1997) Seed dispersers of the monocarpic herbaceous bamboo Pharus virescens (Poaceae: Bambusoideae) found in the Neotropical rain forest of La Macarena and Tinigua National Park of Colombia. Tropics 7:153–159
Janzen DH (1976) Why bamboos wait so long to flower. Ann Rev Ecol Syst 7:347–391
Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125:1198–1205
Kobayashi M (1997) Phylogeny of world bamboos analyzed by restriction fragment length polymorphisms of chloroplast DNA. In: Chapman GP (ed) The bamboos. Academic Press, London, pp 227–236
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kyozuka J, Kobayashi T, Morita M, Shimamoto K (2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41:710–718
McClure FA (1966) The bamboos: a fresh perspective. Harvard University Press, Cambridge
Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007) Evolution and development of inflorescence architectures. Science 316:1452–1455
Raven PH, Axelrod DI (1974) Angiosperm biogeography and past continental movements. Ann Mo Bot Gard 61:539–673
Small RL, Cronn RC, Wendel JF (2004) L. A. S. Johnson review No.2 Use of nuclear genes for phylogeny reconstruction in plants. Aust Syst Bot 17:145–170
Soderstrom TR (1981) Some evolutionary trends in the Bambusoideae (Poaceae). Ann Mo Bot Gard 68:15–47
Soderstrom TR, Ellis RP (1986) The position of bamboo genera and allies in a system of grass classification. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution Press, Washington D. C., pp 225–238
Soderstrom TR, Judziewicz EJ (1987) Systematics of the amphi-Atlantic bambusoid genus Streptogyna (Poaceae). Ann Mo Bot Gard 74:871–888
Soderstrom TR, Ellis RP, Judziewicz EJ (1987) The Phareae and Streprogyneae (Poaceae) of Sri Lanka: a morphological-anatomical study. Smithson Contrib Bot 65:1–27
Swofford DL (2002) PAUP* 4.0 Beta: phylogenetic analysis using parsimony (and other methods). Sinauer, Sunderland
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanimoto T, Kobayashi M (1998) Monocarpic mass flowering of Sasa kurilensis var. jotanii (Bambusoideae) in Mikura-jima, Izu islands, Japan. J Jpn Bot 73:42–47
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Watson L, Dallwitz MJ (1994) The grass genera of the world (revised edition). CABI, Wallingford, pp 683–684
Zhang W, Clark LG (2000) Phylogeny and classification of the Bambusoideae (Poaceae). In: Jacob SWL, Everett J (eds) Grasses: systematic and evolution. CSIRO, Melbourne, pp 35–42
Acknowledgments
We greatly appreciate an anonymous reviewer for the critical comments and invaluable suggestions. This work was partly supported by Grant-in-Aid for Exploratory Research no. 18658062 from the Ministry of Education, Culture, Sports, Science and Technology and Specific Research Assistance B from the Asahi Glass Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10265_2008_181_MOESM1_ESM.xls
S1. The percentages of pairwise divergence in an entire sequence, i.e., in the exons and introns of 27 samples, were calculated as a lower-triangular data matrix. Appendix (XLS 36 kb).
Rights and permissions
About this article
Cite this article
Hisamoto, Y., Kashiwagi, H. & Kobayashi, M. Use of flowering gene FLOWERING LOCUS T (FT) homologs in the phylogenetic analysis of bambusoid and early diverging grasses. J Plant Res 121, 451–461 (2008). https://doi.org/10.1007/s10265-008-0181-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-008-0181-9