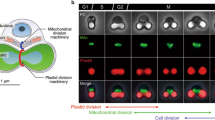
Abstract
Plastids in heterokonts, cryptophytes, haptophytes, dinoflagellates, chlorarachniophytes, euglenoids, and apicomplexan parasites derive from secondary symbiogenesis. These plastids are surrounded by one or two additional membranes covering the plastid-envelope double membranes. Consequently, nuclear-encoded plastid division proteins have to be targeted into the division site through the additional surrounding membranes. Electron microscopic observations suggest that the additional surrounding membranes are severed by mechanisms distinct from those for the division of the plastid envelope. In heterokonts, cryptophytes and haptophytes, the outermost surrounding membrane (epiplastid rough endoplasmic reticulum, EPrER) is studded with cytoplasmic ribosomes and connected to the rER and the outer nuclear envelope. In monoplastidic species belonging to these three groups, the EPrER and the outer nuclear envelope are directly connected to form a sac enclosing the plastid and the nucleus. This nuclear-plastid connection, referred to as the nucleus-plastid consortium (NPC), may be significant to ensure the transmission of the plastids during cell division. The plastid dividing-ring (PD-ring) is a conserved component of the division machinery for both primary and secondary plastids. Also, homologues of the bacterial cell division protein, FtsZ, may be involved in the division of secondary plastids as well as primary plastids, though in secondary plastids they have not yet been localized to the division site. It remains to be examined whether or not dynamin-like proteins and other protein components known to function in the division of primary plastids are used also in secondary plastids. The nearly completed sequencing of the nuclear genome of the diatom Thalassiosira pseudonana will give impetus to molecular and cell biological studies on the division of secondary plastids.

Similar content being viewed by others
References
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Asano T, Yoshioka Y, Kurei S, Sakamoto W, Sodmergen, Machida Y (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38:448–459
Beech PL, Gilson PR (2000) FtsZ and organelle division in protists. Protist 151:11–16
Beech PL, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson PR, McFadden GI (2000) Mitochondrial FtsZ in a chromophyte alga. Science 287:1276–1279
Belcher JH (1969) Some remarks upon Mallomonas papillosa Harris and Bradley and M. calceolus Bradley. Nova Hedwigia 18:257–270
Bhattacharya D, Medlin L (1998) Algal phylogeny and the origin of land plants. Plant Physiol 116:9–15
Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1:298–304
Cavalier-Smith T (1989) The kingdom chromista. In: Green JC, Leadbeater BSC, Diver WL (eds) The chromophyte algae. Oxford University Press, New York, pp 381–407
Cavalier-Smith T (2002a) The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int J Syst Evol Microbiol 52:297–354
Cavalier-Smith T (2002b) Nucleomorphs: enslaved algal nuclei. Curr Opin Microbiol 5:612–619
Cavalier-Smith T (2003) Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Phil Trans R Soc Lond B 358:109–134
Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10:507–516
Douglas SE (1998) Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev 8:655–661
Douglas SE, Zauner S, Fraunholz M, Beaton M, Penny S, Deng L-T, Wu X, Reith M, Cavalier-Smith T, Maier U-G (2001) The highly reduced genome of an enslaved algal nucleus. Nature 410:1091–1096
Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C (1999) Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol 1:239–251
Fraunholz MJ, Moerschel E, Maier UG (1998) The chloroplast division protein FtsZ is encoded by a nucleomorph gene in cryptomonads. Mol Gen Genet 260:207–211
Fulgosi H, Gerdes L, Westphal S, Glockmann C, Soll J (2002) Cell and chloroplast division requires ARTEMIS. Proc Natl Acad Sci USA 99:11501–11506
Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA 100:4328–4333
Gibbs SP (1962) Nuclear envelope-chloroplast relationships in algae. J Cell Sci 14:433–444
Gibbs SP (1978) The chloroplasts of Euglena may have evolved from symbiotic green algae. Can J Bot 56:2883–2889
Gibbs SP (1981) The chloroplast endoplasmic reticulum: structure, function, and evolutionary significance. Int Rev Cytol 72:49–99
Gillott M, Gibbs SP (1980) The cryptomonad nucleomorph: its ultrastructure and evolutionary significance. J Phycol 16:558–568
Gilson PR, McFadden GI (1996) The miniaturized nuclear genome of a eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc Natl Acad Sci USA 93:7737–7742
Gilson PR, McFadden GI (2002) Jam packed genomes—a preliminary, comparative analysis of nucleomorphs. Genetica 115:13–28
Greenwood AD (1974) The cryptophyta in relation to phylogeny and photosynthesis. No. 32. In: Sanders JV, Goodchild DJ (eds) 8th International Conference on Electron Microscopy, Australian Academy of Science, Canberra
Greenwood AD, Griffiths HB, Santore UJ (1977) Chloroplasts and cell compartments in Cryptophyceae. Br Phycol J 12:119
Gu X, Verma DP (1996) Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J 15:695–704
Hales KG, Fuller MT (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90:121–129
Hashimoto H (1986) Double ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 135:166–172
Hashimoto H (1997) Electron-opaque annular structure girdling the constricting isthmus of the dividing chloroplasts of Heterosigma akashiwo (Raphidophyceae, Chromophyta). Protoplasma 197:210–216
Hashimoto H (2003) Plastid division: its origins and evolution. Int Rev Cytol 222:63–98
Hibberd DJ, Norris RE (1984) Cytology and ultrastructure of Chlorarachnion reptans (Chlorarachniophyta division nova, Chlorarachniophyceae classis nova). J Phycol 20:310–330
Hinshaw JE (2000) Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol 16:483–519
van den Hoek C, Mann DG, Jahns HM (1995) Algae: an introduction to phycology. Cambridge University Press, New York
Hong ZL, Geisler-Lee CJ, Zhang ZM, Verma DPS (2003) Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol 53:297–312
Hopkins J, Fowler R, Krishna S, Wilson I, Mitchell G, Bannister L (1999) The plastid in Plasmodium falciparum asexual blood stages: a three-dimensional ultrastructural analysis. Protist 150:283–292
Iino M, Hashimoto H (2003) Intermediate features of cyanelle division of Cyanophora paradoxa (Glaucocystophyta) between cyanobacterial and plastid division. J Phycol 39:561–569
Ishida K, Cavalier-Smith T, Green BR (2000) Endomembrane structure and the chloroplast protein targeting pathway in Heterosigma akashiwo (Raphidophyceae, Chromista). J Phycol 36:1135–1144
Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor MinE plays a role in chloroplast division. Plant Physiol 127:1644–1655
Jin J Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13:1511–1525
Keeling P (2004) A brief history of plastids and their hosts. Protist 155:3–7
Kiefel BR, Gilson PR, Beech PL (2004) Diverse eukaryotes have retained mitochondrial homologues of the bacterial division protein FtsZ. Protist 155:105–115
Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M (2003) Dynamin-like protein 1 is involved in peroxysomal fission. J Biol Chem 278:8597–8605
Kochs G, Haener M, Aebi U, Haller O (2002) Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J Biol Chem 277:14172–14176
Kuroiwa T, Sakai A, Takahashi H, Toda K, Itoh R (1998) The division apparatus of plastid and mitochondria. Int Rev Cytol 181:1–41
Labrousse AM, Zappaterra MD, Rube DA, van Bliek AM (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 4:815–826
Li X, Gould SJ (2003) The dynamin-like GTPase DLP1 is essential for peroxysome division and is recruited to peroxysomes in part by PEX11. J Biol Chem 278:17012–17020
Löffelhardt W, Bohnert HJ, Bryant DA (1997) The cyanelles of Cyanophora paradoxa. Crit Rev Plant Sci 16:393–413
Ludwig M, Gibbs SP (1985) DNA is present in the nucleomorphs of cryptomonads: further evidence that the chloroplast evolved from a eukaryotic endosymbiont. Protoplasma 127:9–20
Ludwig M, Gibbs SP (1989) Evidence that the nucleomorphs of Chlorarachnion reptans (Chlorarachniophyceae) are vestigial nuclei: morphology, division and DNA-DAPI fluorescence. J Phycol 25:385–394
Lutkenhaus J, Addinall SG (1997) Bacterial cell division and the Z ring. Annu Rev Biochem 66:93–116
Maier UG, Douglas SE, Cavalier-Smith T (2000) The nucleomorph genomes of cryptophytes and chlorarachniophytes. Protist 151:103–109
Maple J, Fujiwara MT, Kitahata N, Lawson T, Baker NR, Yoshida S, Moller SG (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabodopsis. Curr Biol 14:776–781
Margolin W (2000) Themes and variations in prokaryotic cell division. FEMS Microbiol Rev 24:531–548
Marks B, Stowel MHB, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT (2001) GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410:231–235
Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima S, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta N, Nishizawa S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol 127:1656–1666
McFadden GI, Gilson PR, Douglas SE (1994) The photosynthetic endosymbiont in cryptomonad cells produces both chloroplast and cytoplasmic-type ribosomes. J Cell Sci 107:649–657
McKerracher L, Gibbs SP (1982) Cell and nucleomorph division in the alga Cryptomonas. Can J Bot 60:2440–2452
Mita T, Kuroiwa T (1988) Division of plastids by a plastid-dividing ring in Cyandium caldarium. Protoplasma Suppl 1:133–152
Mita T, Kanbe T, Tanaka K, Kuroiwa T (1986) A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130:211–213
Miyagishima S, Takahara M, Kuroiwa T (2001a) Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13:707–721
Miyagishima S, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2001b) Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13:2257–2268
Miyagishima S, Nishida K, Kuroiwa T (2003a) An evolutionary puzzle: chloroplast and mitochondrial division rings. Trends Plant Sci 8:432–438
Miyagishima S, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, Kuroiwa T (2003b) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15:655–665
Miyagishima S, Nozaki H, Nishida K, Nishida K, Matsuzaki M, Kuroiwa T (2004) Two types of FtsZ proteins in mitochondrial and red-lineage chloroplasts: the duplication of FtsZ is implicated in endosymbiosis. J Mol Evol 58:291–303
Moestrup Ø (1982) Flagellar structure in algae: a review, with new observations particularly on the Chrysophyceae, Phaeophyceae (Fucophyceae), Euglenophyceae, and Rickertia. Phycologia 21:427–528
Moestrup Ø, Sengco M (2001) Ultrastructural studies on Bigelowiella natans, gen. et sp. nov., a chlorarachniophyte flagellate. J Phycol 37:624–646
Mori T, Kuroiwa H, Takahara M, Miyagishima S, Kuroiwa T (2001) Visualization of an FtsZ-ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol 42:555–559
Morrall S, Greenwood AD (1982) Ultrastructure of nucleomorph division in species of Cryptophyceae and its evolutionary implications. J Cell Sci 54:311–328
Nishida K, Takahara M, Miyagishima S, Kuroiwa H, Matsuzaki M, Kuroiwa T (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA 100:2146–2151
Nozaki H, Matsuzaki M, Takahara M, Misumi O, Kuroiwa H, Hasegawa M, Shin-I T, Kohara Y Ogasawara N, Kuroiwa T (2003) The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondrial-containing eukaryotes and an alternative hypothesis on the origin of plastids. J Mol Evol 56:485–497
Osteryoung KW, McAndrew RS (2001) The plastid division machine. Annu Rev Plant Physiol Plant Mol Biol 52:315–333
Osteryoung KW, Nunnari J (2003) The division of endosymbiotic organelles. Science 302:1698–1704
Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376:473–474
Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10:1991–2004
Palmer JD (2003) The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol 39:4–11
Praefcke GJK, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5:133–147
Pyke KA (1999) Plastid division and development. Plant Cell 11:549–556
Pyke KA, Leech RM (1994) A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol 104:201–207
RayChaudhuri D, Park JT (1992) Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–254
Raynaud C, Cassier-Chauvat C, Perennes C, Bergounioux C (2004) An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16:1801–1811
Robertson EJ, Rutherford SM, Leech RM (1996) Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol 112:149–159
Schmid A-MM (2003) Endobacteria in the diatom Pinnularia (Bacillariophyceae). I. “Scattered ct-nucleoids” explained: DAPI-DNA complexes stem from exoplastidial bacteria boring into the chloroplasts. J Phycol 39:122–138
Schwartzbach SD, Osafune T, Löffelhardt W (1998) Protein import into cyanelles and complex chloroplasts. Plant Mol Biol 38:247–263
Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, Masuda T, Takamiya K, (2004) ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol 45:960–967
Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant molecular gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95:4368–4373
Striepen B, Crawford MJ, Shaw MK, Tilney LG, Seeber F, Roos DS (2000) The plastid of Toxoplasma gondii is divided by association with the centrosomes. J Cell Biol 151:1423–1434
Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Sakai A, Kawano S, Kuroiwa T (1999) Two types of ftsZ genes isolated from the unicellular primitive red alga Galdieria sulphuraria. Plant Cell Physiol 40:784–791
Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Sakai A, Kawano S, Kuroiwa T (2000) A putative mitochondrial ftsZ gene is encoded in the unicellular primitive red alga Cyanidioschyzon merolae. Mol Gen Genet 264:452–460
Taylor FJR (1974) Implications and extensions of the serial endosymbiosis theory of the origin of eukaryotes. Taxon 23:229–258
Vaughan S, Wickstead B, Gull K, Addinall SG (2004) Molecular evolution of FtsZ protein sequences encoded within the genomes of Archea, Bacteria, and Eukaryota. J Mol Evol 58:19–39
Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153:111–119
Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15:1918–1933
Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timing for the origin of photosynthetic eukaryotes. Mol Biol Evol 21:809–818
Zaslavskaia LA, Lippmeier JC, Grossman AR, Apt KE (2000) Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol 36:379–386
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hashimoto, H. The ultrastructural features and division of secondary plastids. J Plant Res 118, 163–172 (2005). https://doi.org/10.1007/s10265-005-0214-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-005-0214-6