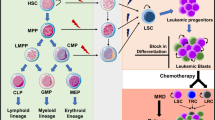
Abstract
Circular RNAs (circRNAs) have been recently identified as important regulators of various diseases, especially cancer. However, the roles of circRNAs in hematologic malignancies have been rarely reported. This study aimed to identify a specific circRNA expression profile in patients with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML), and to evaluate the biological roles of circRNA in MDS and AML for understanding their clinical significance. Reverse transcription-quantitative PCR was performed to validate the expression of circZBTB46. Kruskal–Wallis test, Kaplan–Meier curves, and the Cox regression model were employed to analyze the clinical significance of circZBTB46. Two specific shRNAs as well as an expression lentiviral vector of circZBTB46 were constructed to identify the biological function of circZBTB46. The impact of circZBTB46 on leukemia cell proliferation, cell cycle distribution, and apoptosis was confirmed using cell viability assay and flow cytometry analysis. The expression of circZBTB46 gradually increased in patients with higher-risk MDS and AML, as compared to controls. CircZBTB46 expression was significantly correlated with important clinical parameters of MDS, including WHO classification, absolute neutrophil count (ANC), marrow blast, IPSS karyotype, IPSS/IPSS-R risk groups, and AML transformation. CircZBTB46 expression was also associated with ANC, marrow blast, cytogenetic risk groups, FLT3-ITD mutation, and treatment response in AML patients. Furthermore, circZBTB46 overexpression was significantly correlated with shorter overall survival (OS, P = 0.0342, median survival time 18.5 vs. 45.4 months) and leukemia-free survival (LFS, P = 0.0421) in MDS, also with the shorter OS in AML (P = 0.0293, median survival time 11.6 vs. 16.9 months). Functional studies revealed that silencing circZBTB46 expression significantly inhibited proliferation and induced apoptosis in SKM-1, THP-1, and K562 cell lines, while rescue experiments alleviated the siRNA-mediated growth inhibition and apoptosis in these leukemic cells. The present data suggested the essential oncogenic role of circZBTB46, as a progression and survival indicator in both MDS and AML.




Similar content being viewed by others
Data availability
The microarray data of our study were uploaded into the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/GSE163386). Further inquiries can be directed to the corresponding author.
References
Montalban-Bravo G, Garcia-Manero G. Myelodysplastic syndromes: 2018 update on diagnosis, risk-stratification and management. Am J Hematol. 2018;93:129–47.
de Sousa JC, da Nóbrega IM, Costa MB, et al. Dysregulation of interferon regulatory genes reinforces the concept of chronic immune response in myelodysplastic syndrome pathogenesis. Hematol Oncol. 2019;37:523–6.
Montes P, Bernal M, Campo LN, et al. Tumor genetic alterations and features of the immune microenvironment drive myelodysplastic syndrome escape and progression. Cancer Immunol Immunother. 2019;68:2015–27.
Tazi Y, Arango-Ossa JE, Zhou Y, et al. Unified classification and risk-stratification in Acute Myeloid Leukemia. Nat Commun. 2022;13:4622.
Zhou Q, Shu X, Chai Y, Liu W, Li Z, Xi Y. The non-coding competing endogenous RNAs in acute myeloid leukemia: biological and clinical implications. Biomed Pharmacother. 2023;163:114807.
Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94.
Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869-881.e813.
Long F, Lin Z, Long Q, et al. CircZBTB46 Protects acute myeloid leukemia cells from ferroptotic cell death by upregulating SCD. Cancers (Basel). 2023;15
Chen J, Kao YR, Sun D, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 2019;25:103–10.
Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet. 2016;17:407–21.
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Jamal M, Song T, Chen B, et al. Recent progress on circular RNA research in acute myeloid leukemia. Front Oncol. 2019;9:1108.
Su M, Xiao Y, Ma J, et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90.
Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–42.
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777.
Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152.
Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98.
Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–76.
Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–6.
Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–9.
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11.
Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22-37.e29.
Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–14.
Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9-21.e27.
Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–94.
Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8:1387–402.
Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218.
Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120:e83-92.
Hirsch S, Blätte TJ, Grasedieck S, et al. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica. 2017;102:2039–47.
Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18.
Chen H, Liu T, Liu J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47:1998–2007.
Wu DM, Wen X, Han XR, et al. Role of Circular RNA DLEU2 in Human Acute Myeloid Leukemia. Mol Cell Biol. 2018;38
Yi YY, Yi J, Zhu X, et al. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J Cell Physiol. 2019;234:3711–9.
Xia L, Wu L, Bao J, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2018;503:385–90.
Pan Y, Lou J, Wang H, et al. CircBA9.3 supports the survival of leukaemic cells by up-regulating c-ABL1 or BCR-ABL1 protein levels. Blood Cells Mol Dis. 2018;73:38–44.
Trsova I, Hrustincova A, Krejcik Z, et al. Expression of circular RNAs in myelodysplastic neoplasms and their association with mutations in the splicing factor gene SF3B1. Mol Oncol. 2023
Wedge E, Ahmadov U, Hansen TB, et al. Impact of U2AF1 mutations on circular RNA expression in myelodysplastic neoplasms. Leukemia. 2023;37:1113–25.
Fu Y, He S, Li C, et al. Detailed profiling of m6A modified circRNAs and synergistic effects of circRNA and environmental risk factors for coronary artery disease. Eur J Pharmacol. 2023;951:175761.
Yu J, Yan WZ, Zhang XH, et al. Down-regulation of the Smad signaling by circZBTB46 via the Smad2-PDLIM5 axis to inhibit type I collagen expression. J Geriatr Cardiol. 2023;20:431–47.
Acknowledgements
We would like to thank the patients and physicians enrolled in this study, who came from eight hospitals in the Shanghai Leukemia Cooperative Group.
Funding
This work was supported by the grant from the National Natural Science Foundation of China (NSFC) [grant number 82000134].
Author information
Authors and Affiliations
Contributions
XW and XS designed the project. SL and GZ performed the experiments and drafted the manuscript. SL and GZ contributed equally to this study. WW and NL performed some experiments and analyzed the data. WW and QW collected the patient samples. XS and XW revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests associated with the manuscript.
Ethics approval and consent to participate
The present study, which involved human samples, obtained approval from the Research Ethics Committee of Huashan Hospital, Fudan University. Each patient provided informed consent for participation.
Consent for publication
All authors agree to publish.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, S., Zhao, G., Wu, W. et al. CircZBTB46 predicts poor prognosis and promotes disease progression of myelodysplastic syndromes and acute myeloid leukemia. Clin Exp Med 23, 4649–4664 (2023). https://doi.org/10.1007/s10238-023-01243-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01243-6