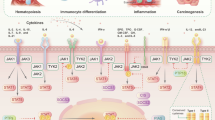
Abstract
Signal transducers and activators of transcription or STAT are proteins that consist of various transcription factors that are responsible for activating genes regarding cell proliferation, differentiation, and apoptosis. They commonly activate several cytokine, growth, or hormone factors via the JAK-STAT signaling pathway by tyrosine phosphorylation which are responsible for giving rise to numerous immune responses. Mutations within the Janus-Kinases (JAKs) or the STATs can set off the commencement of various malfunctions of the immune system of the body; carcinogenesis being an inevitable outcome. STATs are known to act as both oncogenes and tumor suppressor genes which makes it a hot topic of investigation. Various STATs related mechanisms are currently being investigated to analyze its potential of serving as a therapeutic base for numerous immune diseases and cancer; a deeper understanding of the molecular mechanisms involved in the signaling pathways can contribute to the same. This review will throw light upon each STAT member in causing cancer malignancies by affecting subsequent signaling pathways and its genetic and epigenetic associations as well as various inhibitors that could be used to target these pathways thereby devising new treatment options. The review will also focus upon the therapeutic advances made in cancers that most commonly affect people and discuss how STAT genes are identified as prognostic markers.







Similar content being viewed by others
Abbreviations
- STAT: :
-
Signal transducers and activators of transcription
- JAK: :
-
Janus kinase
- IFN: :
-
Interferon
- DN: :
-
Dominant negative
- EGF: :
-
Epidermal growth factor
- PDGF: :
-
Platelet-derived growth factor
- CSF-1::
-
Colony-stimulating factor 1
- IL: :
-
Interleukin
- BCL: :
-
B-cell lymphoma
- TRADD::
-
Tumor necrosis factor receptor 1-associated death domain protein
- Mdm2::
-
Murine double minute 2
- c-Myc: :
-
Cellular myelocytomatosis oncogene
- MHC: :
-
Major histocompatibility complex
- bFGF: :
-
Basic fibroblast growth factor
- PTEN/Akt: :
-
Phosphatase and tensin homolog deleted on chromosome 10/Ak strain transforming
- HNSCC: :
-
head and neck squamous cell carcinoma
- TRAIL: :
-
TNF-related apoptosis-inducing ligand
- Fra-1: :
-
Fos-related antigen 1
- TF: :
-
Transcription factor
- FBXW7: :
-
F-box/WD repeat-containing protein 7
- EGFR: :
-
Epidermal growth factor receptor
- ISGF3: :
-
Interferon stimulated gene factor 3
- VEGF: :
-
Vascular endothelial growth factor
- MMP 2: :
-
Matrix metalloproteinase-2
- HFIH: :
-
Hexane fraction of I. helenium
- SH2: :
-
Src Homology 2
- HBV: :
-
Hepatitis B virus
- HCC: :
-
Hepatocellular carcinoma
- DC: :
-
Dendritic cells
- NOD: :
-
Nonobese diabetic
- BM: :
-
Bone marrow
- GM-CSF: :
-
Granulocyte–macrophage colony-stimulating factor
- Tyk: :
-
Tyrosine kinase
- SOCS: :
-
Suppressors of cytokine signaling proteins
- CIS: :
-
Cytokine inhibitor Src homology 2-containing protein
- PIAS: :
-
Protein inhibitors of activated STATs
- ABC: :
-
ATP binding cassette
- MCl -1: :
-
Myeloid cell leukemia 1
- CML: :
-
Chronic myelogenous leukemia
- AML: :
-
Acute myelogenous leukemia
- FLT3-IND: :
-
FLT3 internal tandem duplication
- MSM: :
-
Methylsufonylmethane
- HER: :
-
Human epidermal growth factor receptor
- ERK: :
-
Extracellular signal-regulated kinase
- Th: :
-
T helper
- IAP: :
-
Inhibitors of apoptosis proteins
- Cdk: :
-
Cyclin-dependent kinases
- DNMT: :
-
DNA methyltransferases
- PCa: :
-
Prostate cancer
- ADT: :
-
Androgen deprivation therapy
- BRCA1: :
-
Breast Cancer gene 1
- AR: :
-
Androgen receptor
- ATF 3: :
-
Activating transcription factor 3
- NSCLC: :
-
Non small cell lung cancer
- CIN: :
-
Cervical intraepithelial neoplasia
- TNBC: :
-
Triple negative breast cancer
- OC: :
-
Ovarian cancer
- CXCL: :
-
Chemokine ligand
- EOC: :
-
Epithelial ovarian cancer
- JNK: :
-
Jun N-terminal kinase
- PAK: :
-
Serine/threonine-protein kinase
- PPAR: :
-
Peroxisome proliferator-activated receptor
- CRC: :
-
Colorectal cancers
- COX: :
-
Cyclooxygenase
- IL22RA1: :
-
Interleukin 22 receptor, alpha 1
- OS: :
-
Overall survival
- PDK4: :
-
Pyruvate Dehydrogenase Kinase 4
- TCA:
-
Tricarboxylic acid
- RAS:
-
Rat sarcoma
- MAPK:
-
Mitogen-activated protein kinase
- PI3K:
-
Phosphatidylinositol-3-kinase
- AKT:
-
Protein kinase B
References
Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84(3):331–4. https://doi.org/10.1016/s0092-8674(00)81277-5.
Loh CY, Arya A, Naema AF, Wong WF, Sethi G, Looi CY. Signal transducer and activator of transcription (stats) proteins in cancer and inflammation: functions and therapeutic implication. Front Oncol. 2019;21(9):48.
Benekli M, Baer MR, Baumann H, Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101(8):2940–54. https://doi.org/10.1182/blood-2002-04-1204.
Meissl K, Macho-Maschler S, Müller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20.
Hsu KS, Zhao X, Cheng X, et al. Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon α-mediated inhibition of angiogenesis. J Biol Chem. 2017;292(24):10048–60.
Huang S, Bucana CD, Van Arsdall M, Fidler IJ. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene. 2002;21(16):2504–12.
Zhang X, Li X, Tan F, Yu N, Pei H. STAT1 Inhibits MiR-181a expression to suppress colorectal cancer cell proliferation through PTEN/Akt. J Cell Biochem. 2017;118(10):3435–43.
Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24(130):19–29.
Xi S, Dyer KF, Kimak M, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98(3):181–9.
Pitroda SP, Wakim BT, Sood RF, et al. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 2009;5(7):68.
Zhu H, Wang Z, Xu Q, et al. Inhibition of STAT1 sensitizes renal cell carcinoma cells to radiotherapy and chemotherapy. Cancer Biol Ther. 2012;13(6):401–7.
Roberts D, Schick J, Conway S, et al. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer. 2005;92(6):1149–58.
Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci U S A. 2004;101(6):1714–9.
Choi EA, Lei H, Maron DJ, et al. Stat1-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand and the cell-surface death signaling pathway by interferon beta in human cancer cells. Cancer Res. 2003;63(17):5299–307.
Zhang M, Liang L, He J, et al. Fra-1 inhibits cell growth and the warburg effect in cervical cancer cells via stat1 regulation of the p53 signaling pathway. Front Cell Dev Biol. 2020;30(8):579629.
Ry FZ, Farrar JD. STAT2: A shape-shifting anti-viral super STAT. JAKSTAT. 2013;2(1):e23633.
Verhoeven Y, Tilborghs S, Jacobs J, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol. 2020;60:41–56. https://doi.org/10.1016/j.semcancer.2019.10.002.
Steen HC, Gamero AM. STAT2 phosphorylation and signaling. JAKSTAT. 2013;2(4):e25790.
Lee CJ, An HJ, Cho ES, et al. Stat2 stability regulation: an intersection between immunity and carcinogenesis. Exp Mol Med. 2020;52(9):1526–36. https://doi.org/10.1038/s12276-020-00506-6.
Ebersbach C, Beier AK, Thomas C, Erb HHH. Impact of STAT proteins in tumor progress and therapy resistance in advanced and metastasized prostate cancer. Cancers (Basel). 2021;13(19):4854.
Chen X, Huang J, Lü Y. High expression of STAT2 in ovarian cancer and its effect on metastasis of ovarian cancer cells. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40(1):34–41.
Liang Z, Gao LH, Cao LJ, et al. Detection of STAT2 in early stage of cervical premalignancy and in cervical cancer. Asian Pac J Trop Med. 2012;5(9):738–42.
Gamero AM, Young MR, Mentor-Marcel R, et al. STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev Res (Phila). 2010;3(4):495–504.
Furth PA. STAT signaling in different breast cancer sub-types. Mol Cell Endocrinol. 2014;382(1):612–5.
Uluer ET, Aydemir I, Inan S, Ozbilgin K, Vatansever HS. Effects of 5-fluorouracil and gemcitabine on a breast cancer cell line (MCF-7) via the JAK/STAT pathway. Acta Histochem. 2012;114(7):641–6.
Schaber JD, Fang H, Xu J, Grimley PM, Rui H. Prolactin activates Stat1 but does not antagonize Stat1 activation and growth inhibition by type I interferons in human breast cancer cells. Cancer Res. 1998;58(9):1914–9.
Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13(2):211–7.
Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. 2013;2013:421821.
Kanda N, Seno H, Konda Y, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23(28):4921–9.
Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. Prostate. 2002;51(4):241–6.
Xie TX, Huang FJ, Aldape KD, et al. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66(6):3188–96.
Ma JH, Qin L, Li X. Role of STAT3 signaling pathway in breast cancer. Cell Commun Signal. 2020;18(1):33.
Wang G, Chen JH, Qiang Y, Wang DZ, Chen Z. Decreased STAT4 indicates poor prognosis and enhanced cell proliferation in hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3983–93.
Verhoeven Y, Tilborghs S, Jacobs J, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol. 2020;60:41–56.
Ronald Hoffman EJ, Leslie B, Silberstein E, et al. Hematology. 7th ed. Newyork: Elsevier; 2018. p. 163–75.
Yang Z, Chen M, Fialkow LB, Ellett JD, Wu R, Nadler JL. Inhibition of STAT4 activation by lisofylline is associated with the protection of autoimmune diabetes. Ann N Y Acad Sci. 2003;1005:409–11.
Chiang PH, Wang L, Bonham CA, et al. Mechanistic insights into impaired dendritic cell function by rapamycin: inhibition of Jak2/Stat4 signaling pathway. J Immunol. 2004;172(3):1355–63.
Lin JT, Martin SL, Xia L, Gorham JD. TGF-beta 1 uses distinct mechanisms to inhibit IFN-gamma expression in CD4+ T cells at priming and at recall: differential involvement of Stat4 and T-bet. J Immunol. 2005;174(10):5950–8.
Kornfeld JW, Grebien F, Kerenyi MA, et al. The different functions of Stat5 and chromatin alteration through Stat5 proteins. Front Biosci. 2008;1(13):6237–54.
Walker SR, Xiang M, Frank DA. Distinct roles of STAT3 and STAT5 in the pathogenesis and targeted therapy of breast cancer. Mol Cell Endocrinol. 2014;382(1):616–21.
Igelmann S, Neubauer HA, Ferbeyre G. STAT3 and STAT5 Activation in Solid Cancers. Cancers (Basel). 2019;11(10):1428.
Schepers H, van Gosliga D, Wierenga AT, Eggen BJ, Schuringa JJ, Vellenga E. STAT5 is required for long-term maintenance of normal and leukemic human stem/progenitor cells. Blood. 2007;110(8):2880–8.
Talati PG, Gu L, Ellsworth EM, et al. Jak2-Stat5a/b Signaling Induces Epithelial-to-Mesenchymal Transition and Stem-Like Cell Properties in Prostate Cancer. Am J thol. 2015;185(9):2505–22.
Sumiyoshi H, Matsushita A, Nakamura Y, et al. Suppression of STAT5b in pancreatic cancer cells leads to attenuated gemcitabine chemoresistance, adhesion and invasion. Oncol Rep. 2016;35(6):3216–26.
Kollmann S, Grundschober E, Maurer B, et al. Twins with different personalities: STAT5B-but not STAT5A-has a key role in BCR/ABL-induced leukemia. Leukemia. 2019;33(7):1583–97. https://doi.org/10.1038/s41375-018-0369-5.
Li Z, Chen C, Chen L, et al. STAT5a confers doxorubicin resistance to breast cancer by regulating ABCB1. Front Oncol. 2021;15(11):697950.
Maurer B, Kollmann S, Pickem J, Hoelbl-Kovacic A, Sexl V. STAT5A and STAT5B-Twins with Different Personalities in Hematopoiesis and Leukemia. Cancers (Basel). 2019;11(11):1726.
Nevalainen MT, Xie J, Torhorst J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22(11):2053–60.
Nelson EA, Walker SR, Weisberg E, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117(12):3421–9.
Nelson EA, Walker SR, Xiang M, et al. The STAT5 inhibitor pimozide displays efficacy in models of acute myelogenous leukemia driven by FLT3 mutations. Genes Cancer. 2012;3(7–8):503–11.
Gallipoli P, Cook A, Rhodes S, et al. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of CML CD34+ cells in vitro and in vivo. Blood. 2014;124(9):1492–501.
Lim EJ, Hong DY, Park JH, et al. Methylsulfonylmethane suppresses breast cancer growth by down-regulating STAT3 and STAT5b pathways. PLoS One. 2012;7(4):e33361.
Walker SR, Nelson EA, Zou L, et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7(6):966–76. https://doi.org/10.1158/1541-7786.MCR-08-0238.
Lee H, Kim JW, Choi DK, et al. Poziotinib suppresses ovarian cancer stem cell growth via inhibition of HER4-mediated STAT5 pathway. Biochem Biophys Res Commun. 2020;526(1):158–64. https://doi.org/10.1016/j.bbrc.2020.03.046.
Hideho Okada, Jacques Banchereau, Michael T Lotze. CHAPTER 10 - Interleukin-4, In:The Cytokine Handbook (Fourth Edition),Editor(s): Angus W. Thomson, Michael T. Lotze, Academic Press,2003,Pages 227–262,ISBN 9780126896633
My Cancer Genome Genetically Informed Cancer Medicine; Available from: https://www.mycancergenome.org/content/gene/stat6/
Delgado-Ramirez Y, Colly V, Gonzalez GV, Leon-Cabrera S. Signal transducer and activator of transcription 6 as a target in colon cancer therapy. Oncol Lett. 2020;20(1):455–64. https://doi.org/10.3892/ol.2020.11614.
Binnemars-Postma K, Bansal R, Storm G, Prakash J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018;32(2):969–78. https://doi.org/10.1096/fj.201700629R.
Fu C, Jiang L, Hao S, et al. Activation of the IL-4/STAT6 signaling pathway promotes lung cancer progression by increasing M2 myeloid cells. Front Immunol. 2019;10:2638. https://doi.org/10.3389/fimmu.2019.02638.
Clevenger CV. Roles and regulation of stat family transcription factors in human breast cancer. Am J Pathol. 2004;165(5):1449–60. https://doi.org/10.1016/S0002-9440(10)63403-7.
Alvarez JV, Frank DA. Genome-wide analysis of STAT target genes: elucidating the mechanism of STAT-mediated oncogenesis. Cancer Biol Ther. 2004;3(11):1045–50. https://doi.org/10.4161/cbt.3.11.1172.
Seghal PD. Epigenetic regulation of transcription factor STAT3 activity in cancer cells. Contemp Oncol/Współczesna Onkol. 2006;10(8):373–7.
Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108(3):1058–64. https://doi.org/10.1182/blood-2005-08-007377.
Wingelhofer B, Neubauer HA, Valent P, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32(8):1713–26. https://doi.org/10.1038/s41375-018-0117-x.
Leslie SW, Soon-Sutton TL, R I A, et al. Prostate Cancer. [Updated 2022 Nov 28]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK470550/
Ebersbach C, Beier AK, Thomas C, Erb HHH. Impact of STAT proteins in tumor progress and therapy resistance in advanced and metastasized prostate cancer. Cancers (Basel). 2021;13(19):4854. https://doi.org/10.3390/cancers13194854.
El-Habr EA, Levidou G, Trigka EA, et al. Complex interactions between the components of the PI3K/AKT/mTOR pathway, and with components of MAPK, JAK/STAT and Notch-1 pathways, indicate their involvement in meningioma development. Virchows Arch. 2014;465(4):473–85. https://doi.org/10.1007/s00428-014-1641-3.
da Silva HB, Amaral EP, Nolasco EL, et al. Dissecting major signaling pathways throughout the development of prostate cancer. Prostate Cancer. 2013;2013:920612. https://doi.org/10.1155/2013/920612.
Dutta P, Sabri N, Li J, Li WX. Role of STAT3 in lung cancer. JAKSTAT. 2015;3(4):e999503. https://doi.org/10.1080/21623996.2014.999503.
Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. https://doi.org/10.1016/s0092-8674(00)81959-5.
Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2023. https://doi.org/10.1038/sj.onc.1206479.
Cheng F, Wang HW, Cuenca A, et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19(3):425–36. https://doi.org/10.1016/s1074-7613(03)00232-2.
Fowler JR, Maani EV, Dunton CJ, Jack BW. Cervical Cancer. Treasure Island (FL): StatPearls Publishing; 2022.
Gutiérrez-Hoya A, Soto-Cruz I. Role of the JAK/STAT pathway in cervical cancer: its relationship with HPV E6/E7 oncoproteins. Cells. 2020;9(10):2297. https://doi.org/10.3390/cells9102297.
Liang Z, Gao LH, Cao LJ, et al. Detection of STAT2 in early stage of cervical premalignancy and in cervical cancer. Asian Pac J Trop Med. 2012;5(9):738–42. https://doi.org/10.1016/S1995-7645(12)60117-5.
Sun CY, Nie J, Huang JP, Zheng GJ, Feng B. Targeting STAT3 inhibition to reverse cisplatin resistance. Biomed Pharmacother. 2019;117:109135. https://doi.org/10.1016/j.canlet.2014.05.005.
Huang LL, Rao W. SiRNA interfering STAT3 enhances DDP sensitivity in cervical cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(13):4098–106.
Uluer ET, Aydemir I, Inan S, Ozbilgin K, Vatansever HS. Effects of 5-fluorouracil and gemcitabine on a breast cancer cell line (MCF-7) via the JAK/STAT pathway. Acta Histochem. 2012;114(7):641–6. https://doi.org/10.1016/j.acthis.2011.11.010.
Furth PA. STAT signaling in different breast cancer sub-types. Mol Cell Endocrinol. 2014;382(1):612–5. https://doi.org/10.1016/j.mce.2013.03.023.
Zhang WJ, Li BH, Yang XZ, et al. IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells. Cytokine. 2008;42(1):39–47. https://doi.org/10.1016/j.cyto.2008.01.016.
Liang R, Chen X, Chen L, et al. STAT3 signaling in ovarian cancer: a potential therapeutic target. J Cancer. 2020;11(4):837–48. https://doi.org/10.7150/jca.35011.
Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res. 2011;317(5):620–31. https://doi.org/10.1016/j.yexcr.2010.12.017.
Wu CJ, Sundararajan V, Sheu BC, Huang RY, Wei LH. Activation of STAT3 and STAT5 signaling in epithelial ovarian cancer progression: mechanism and therapeutic opportunity. Cancers (Basel). 2019;12(1):24. https://doi.org/10.3390/cancers12010024.
Rezaeeyan H, Hassani SN, Barati M, et al. PD-1/PD-L1 as a prognostic factor in leukemia. J Hematopathol. 2017;10:17–24. https://doi.org/10.1007/s12308-017-0293-z.
Shahjahani M, Abroun A, Saki N, Bagher Mohammadi SM, Rezaeeyan H. STAT5: from pathogenesis mechanism to therapeutic approach in acute leukemia. Lab Med. 2020;51(4):345–51. https://doi.org/10.1093/labmed/lmz074.
Zhu CQ, Tsao MS. Prognostic markers in lung cancer: is it ready for prime time? Transl Lung Cancer Res. 2014;3(3):149–58. https://doi.org/10.3978/j.issn.2218-6751.2014.06.09.
Li S, Sheng B, Zhao M, Shen Q, Zhu H, Zhu X. The prognostic values of signal transducers activators of transcription family in ovarian cancer. 2017. Biosci Rep. https://doi.org/10.1042/BSR20170650.
Dong Z, Chen Y, Yang C, et al. STAT gene family mRNA expression and prognostic value in hepatocellular carcinoma. Onco Targets Ther. 2019;3(12):7175–91. https://doi.org/10.2147/OTT.S202122.
Zhang J, Wang F, Liu F, Xu G. Predicting STAT1 as a prognostic marker in patients with solid cancer. Ther Adv Med Oncol. 2020;12:1758835920917558. https://doi.org/10.1177/1758835920917558.
Gordziel C, Bratsch J, Moriggl R, Knösel T, Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br J Cancer. 2013;109(1):138–46. https://doi.org/10.1038/bjc.2013.274.
Klupp F, Diers J, Kahlert C, et al. Expressional STAT3/STAT5 ratio is an independent prognostic marker in colon carcinoma. Ann Surg Oncol. 2015;22(Suppl 3):S1548–55. https://doi.org/10.1245/s10434-015-4485-4.
Liu X, He Z, Li CH, Huang G, Ding C, Liu H. Correlation analysis of JAK-STAT pathway components on prognosis of patients with prostate cancer. Pathol Oncol Res. 2012;18(1):17–23. https://doi.org/10.1007/s12253-011-9410-y.
Pang C, Gu Y, Ding Y, et al. Several genes involved in the JAK-STAT pathway may act as prognostic markers in pancreatic cancer identified by microarray data analysis. Medicine (Baltimore). 2018;97(50):e13297. https://doi.org/10.1097/MD.0000000000013297.
Tu Y, Zhong Y, Fu J, et al. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Med Oncol. 2011;28(1):15–23. https://doi.org/10.1007/s12032-010-9435-1.
Oberhuber M, Pecoraro M, Rusz M, et al. STAT3-dependent analysis reveals PDK4 as independent predictor of recurrence in prostate cancer. Mol Syst Biol. 2020;16(4):e9247.
Chen HH, Chou CY, Wu YH, et al. Constitutive STAT5 activation correlates with better survival in cervical cancer patients treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2012;82(2):658–66. https://doi.org/10.1016/j.ijrobp.2010.11.043.
Shi S, Ma HY, Zhang ZG. Clinicopathological and prognostic value of STAT3/p-STAT3 in cervical cancer: a meta and bioinformatics analysis. Pathol Res Pract. 2021;227:153624. https://doi.org/10.1016/j.prp.2021.153624.
Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. https://doi.org/10.1038/nrc1275.
Guanizo AC, Fernando CD, Garama DJ, Gough DJ. STAT3: a multifaceted oncoprotein. Growth Factors. 2018;36(1–2):1–14.
Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109(9):1139–42.
Furqan M, Akinleye A, Mukhi N, Mittal V, Chen Y, Liu D. STAT inhibitors for cancer therapy. J Hematol Oncol. 2013;6:90. https://doi.org/10.1186/1756-8722-6-90.
Acknowledgements
We thank Sir C.V. Raman Research Park and Department of Biotechnology of SRM Institute of Science and Technology, Kattankulathur, Chennai, India for providing laboratory space.
Funding
This research was supported by the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Govt. of India (Sanction Order No. ECR/2016/000965).
Author information
Authors and Affiliations
Contributions
KS contributed to Conceptualization, supervision and funding acquisition; SB, NJ, SK, RR, VT contributed to investigation, data curation, and writing of the original draft; SB, NJ, SK contributed to visualization; GS, SR, KS contributed to reviewing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jill, N., Bhootra, S., Kannanthodi, S. et al. Interplay between signal transducers and activators of transcription (STAT) proteins and cancer: involvement, therapeutic and prognostic perspective. Clin Exp Med 23, 4323–4339 (2023). https://doi.org/10.1007/s10238-023-01198-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01198-8