Abstract
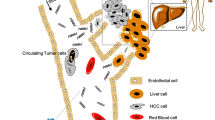
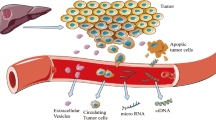
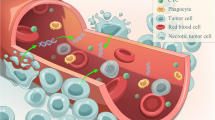
The growing role of precision medicine in hepatocellular carcinoma (HCC) is expected to ameliorate the poor prognosis and high mortality of this highly malignant disease; however, it is faced with challenges such as the low frequency of tissue biopsy. Hence, attention is turning to the circulating tumor DNA (ctDNA), an important component of liquid biopsy. Obtaining molecular information about cancer from blood provides a good prospect in precision oncology including molecular diagnosis, molecular classification, targeted therapy, personalized decision making, and detection of drug-resistance mutations. However, inherent constraints of HCC and ctDNA (like background chronic liver diseases (CLD) and low concentration of ctDNA) along with some technical issues should be well handled and solved before the potential of ctDNA in precision medicine of HCC can be truly realized. In this review, we will focus on the prospects and challenges of ctDNA in HCC precision medicine.

Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Ng CKY, Di Costanzo GG, Terracciano LM, Piscuoglio S. Circulating cell-free DNA in hepatocellular carcinoma: current insights and outlook. Front Med. 2018;5:78.
Li J, Han X, Yu X, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37(1):213.
Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Discov. 2019;18(1):13–4.
Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88.
Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149(5):1226–39.e1224.
Chan SL, Wong AM, Lee K, Wong N, Chan AK. Personalized therapy for hepatocellular carcinoma: Where are we now? Cancer Treat Rev. 2016;45:77–86.
Qu C, Wang Y, Wang P, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci USA. 2019;116(13):6308–12.
Marquardt JU, Thorgeirsson SS. SnapShot: hepatocellular carcinoma. Cancer Cell. 2014;25(4):550.e551.
Zhang Z, Chen P, Xie H, Cao P. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: a systematic review and meta-analysis. Cancer Med. 2020;9(4):1349–64.
Oussalah A, Rischer S, Bensenane M, et al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–47.
Wei L, Huang Y, Zhao R, et al. Detection of promoter methylation status of suppressor of cytokine signaling 3 (SOCS3) in tissue and plasma from Chinese patients with different hepatic diseases. Clin Exp Med. 2018;18(1):79–877.
Zhang YJ, Wu HC, Shen J, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13(8):2378–84.
Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–61.
Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616.
Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12(8):436.
Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30(1):35–51.
Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–888.
Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–92.
Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology (Baltimore, MD). 2007;45(1):42–52.
Lachenmayer A, Alsinet C, Savic R, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18(18):4997–5007.
Wang K, Lim HY, Shi S, et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology (Baltimore, MD). 2013;58(2):706–17.
Toffanin S, Hoshida Y, Lachenmayer A, et al. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140(5):1618–28.e1616.
Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology (Baltimore, MD). 2015;61(6):1945–56.
Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140(5):1501–12.e1502.
Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol. 2018;15(3):183–92.
Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67(12):2204–12.
Ikeda S, Lim JS, Kurzrock R. Analysis of tissue and circulating tumor DNA by next-generation sequencing of hepatocellular carcinoma: implications for targeted therapeutics. Mol Cancer Ther. 2018;17(5):1114–22.
Ikeda S, Tsigelny IF, Skjevik AA, et al. Next-generation sequencing of circulating tumor DNA reveals frequent alterations in advanced hepatocellular carcinoma. Oncologist. 2018;23(5):586–93.
He G, Chen Y, Zhu C, et al. Application of plasma circulating cell-free DNA detection to the molecular diagnosis of hepatocellular carcinoma. Am J Transl Res. 2019;11(3):1428–45.
Huang A, Zhang X, Zhou SL, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer. 2016;7(13):1907–14.
Huang X, Wu C, Fu Y, Guo L, Kong X, Cai H. Methylation analysis for multiple gene promoters in non-small cell lung cancers in high indoor air pollution region in China. Bull Cancer. 2018;105(9):746–54.
Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid biopsies for cancer: coming to a patient near you. J Clin Med. 2017;6(1):3.
Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–43.
Zhang X, Ju S, Wang X, Cong H. Advances in liquid biopsy using circulating tumor cells and circulating cell-free tumor DNA for detection and monitoring of breast cancer. Clin Exp Med. 2019;19(3):271–9.
Su YH, Kim AK, Jain S. Liquid biopsies for hepatocellular carcinoma. Transl Res. 2018;201:84–97.
Husain H, Velculescu VE. Cancer DNA in the circulation: the liquid biopsy. JAMA. 2017;318(13):1272–4.
Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321(14):1391–9.
Lim HY, Merle P, Weiss KH, et al. Phase II studies with refametinib or refametinib plus sorafenib in patients with RAS-mutated hepatocellular carcinoma. Clin Cancer Res. 2018;24(19):4650–61.
Husain H, Melnikova VO, Kosco K, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res. 2017;23(16):4716–23.
Cai ZX, Chen G, Zeng YY, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer. 2017;141(5):977–85.
Cai Z, Chen G, Zeng Y, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res. 2019;25(17):5284–94.
An Y, Guan Y, Xu Y, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res. 2019;11(10):6462–74.
Liao W, Yang H, Xu H, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481–90.
Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci USA. 2016;113(21):6005–100.
Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine. 2016;10:117–23.
Blokzijl F, de Ligt J, Jager M, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538(7624):260–4.
Tang JC, Feng YL, Guo T, Xie AY, Cai XJ. Circulating tumor DNA in hepatocellular carcinoma: trends and challenges. Cell Biosci. 2016;6:32.
Ringelhan M, Pfister D, O'Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(3):222–32.
Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology (Baltimore, MD). 2014;60(6):1983–92.
Pilati C, Letouze E, Nault JC, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25(4):428–41.
Szymanska K, Lesi OA, Kirk GD, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia. West Africa Int J Cancer. 2004;110(3):374–9.
Li X, Wang H, Li T, et al. Circulating tumor DNA/circulating tumor cells and the applicability in different causes induced hepatocellular carcinoma. Curr Probl Cancer. 2019;2019:100516.
Kaseb AO, Sanchez NS, Sen S, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res. 2019;25(20):6107–18.
Marchio A, Amougou Atsama M, Bere A, et al. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med. 2018;18(3):421–31.
Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–51.
Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54.
Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):2415.
Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA. 2015;112(11):E1317–E13251325.
Jiang P, Sun K, Tong YK, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci USA. 2018;115(46):E10925–E1093310933.
Evans JP, Powell BC, Berg JS. Finding the rare pathogenic variants in a human genome. JAMA. 2017;317(18):1904–5.
Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–55.
Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–705.
Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2018;4(6):868–70.
Teutsch SM, Bradley LA, Palomaki GE, et al. The evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP working group. Genet Med. 2009;11(1):3–14.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81472672), International Science and Technology Cooperation Project of Shanghai (18410721900), and Leading Talent Program of Minhang District (2017).
Author information
Authors and Affiliations
Contributions
NR had the idea for the article. JLW performed the literature search and data analysis. JLW, MA, and CHZ wrote the manuscript. JLW designed the tables and figures. NR critically revised the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Weng, J., Atyah, M., Zhou, C. et al. Prospects and challenges of circulating tumor DNA in precision medicine of hepatocellular carcinoma. Clin Exp Med 20, 329–337 (2020). https://doi.org/10.1007/s10238-020-00620-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-020-00620-9