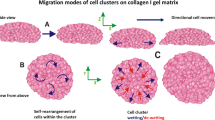
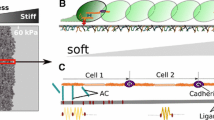
Abstract
Cells migrating in clusters play a significant role in a number of biological processes such as embryogenesis, wound healing, and tumor metastasis during cancer progression. A variety of environmental and biochemical factors can influence the collective migration of cells with differing degrees of cell autonomy and inter-cellular coupling strength. For example, weakly coupled cells can move collectively under the influence of contact guidance from neighboring cells or the environment. Alternatively strongly coupled cells might follow one or more leader cells to move as a single cohesive unit. Additionally, chemical and mechanical signaling between these cells may alter the degree of coupling and determine effective cluster sizes. Being able to understand this collective cell migration process is critical in the prediction and manipulation of outcomes of key biological processes. Here we focus on understanding how various environmental and cellular factors influence small clusters of cells migrating collectively within a 3D fibrous matrix. We combine existing knowledge of single-cell migration in 2D and 3D environments, prior experimental observations of cell–cell interactions and collective migration, and a newly developed stochastic model of cell migration in 3D matrices, to simulate the migration of cell clusters in different physiologically relevant environments. Our results show that based on the extracellular environment and the strength of cell–cell mechanical coupling, two distinct optimal approaches to driving collective cell migration emerge. The ability to effectively employ these two distinct migration strategies might be critical for cells to collectively migrate through the heterogeneous tissue environments within the body.






Similar content being viewed by others
References
Abraham VC, Krishnamurthi V, Taylor DL, Lanni F (1999) The actin-based nanomachine at the leading edge of migrating cells. Biophys J 77(3):1721–1732
Ahmadzadeh H, Webster MR, Behera R, Jimenez Valencia AM, Wirtz D, Weeraratna AT, Shenoy VB (2017) Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proc Natl Acad Sci U S A 114(9):E1617–E1626. https://doi.org/10.1073/pnas.1617037114
Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P (2008) Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem Cell Biol 130(6):1147–54. https://doi.org/10.1007/s00418-008-0529-1
An J, Enomoto A, Weng L, Kato T, Iwakoshi A, Ushida K, Maeda K, Ishida-Takagishi M, Ishii G, Ming S, Sun T, Takahashi M (2013) Significance of cancer-associated fibroblasts in the regulation of gene expression in the leading cells of invasive lung cancer. J Cancer Res Clin Oncol 139(3):379–388. https://doi.org/10.1007/s00432-012-1328-6
Ananthakrishnan R, Ehrlicher A (2007) The forces behind cell movement. Int J Biol Sci 3(5):303
Arima S, Nishiyama K, Ko T, Arima Y, Hakozaki Y, Sugihara K, Koseki H, Uchijima Y, Kurihara Y, Kurihara H (2011) Angiogenic morphogenesis driven by dynamic and heterogeneous collective endothelial cell movement. Development 138(21):4763–76. https://doi.org/10.1242/dev.068023
Arrieumerlou C, Meyer T (2005) A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell 8(2):215–227. https://doi.org/10.1016/j.devcel.2004.12.007
Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rørth P (2007) Two distinct modes of guidance signalling during collective migration of border cells. Nature 448(7151):362–365. https://doi.org/10.1038/nature05965
Bosgraaf L, Van Haastert PJM (2009) The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS ONE 4(4):e5253. https://doi.org/10.1371/journal.pone.0005253
Bronsert P, Enderle-Ammour K, Bader M, Timme S, Kuehs M, Csanadi A, Kayser G, Kohler I, Bausch D, Hoeppner J, Hopt U, Keck T, Stickeler E, Passlick B, Schilling O, Reiss C, Vashist Y, Brabletz T, Berger J, Lotz J, Olesch J, Werner M, Wellner U (2014) Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface: 3d cancer-host interface. J Pathol 234(3):410–422. https://doi.org/10.1002/path.4416
Bruinsma R (2005) Theory of force regulation by nascent adhesion sites. Biophys J 89(1):87–94
Buhl J, Sumpter DJT, Couzin ID, Hale JJ, Despland E, Miller ER, Simpson SJ (2006) From disorder to order in marching locusts. Science 312(5778):1402–6. https://doi.org/10.1126/science.1125142
Burgess BT, Myles JL, Dickinson RB (2000) Quantitative analysis of adhesion-mediated cell migration in three-dimensional gels of RGD-grafted collagen. Ann Biomed Eng 28(1):110–118
Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer J, Danuser G, Montell D (2014) Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157(5):1146–1159. https://doi.org/10.1016/j.cell.2014.03.045
Cai D, Dai W, Prasad M, Luo J, Gov NS, Montell DJ (2016) Modeling and analysis of collective cell migration in an in vivo three-dimensional environment. Proc Natl Acad Sci USA 113(15):E2134. https://doi.org/10.1073/pnas.1522656113
Campbell JJ, Husmann A, Hume RD, Watson CJ, Cameron RE (2017) Development of three-dimensional collagen scaffolds with controlled architecture for cell migration studies using breast cancer cell lines. Biomaterials 114:34–43. https://doi.org/10.1016/j.biomaterials.2016.10.048
Carey SP, Kraning-Rush CM, Williams RM, Reinhart-King CA (2012) Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials 33(16):4157–65. https://doi.org/10.1016/j.biomaterials.2012.02.029
Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R (2008) Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456(7224):957–961. https://doi.org/10.1038/nature07441
Cavagna A, Cimarelli A, Giardina I, Parisi G, Santagati R, Stefanini F, Viale M (2010) Scale-free correlations in starling flocks. Proc Natl Acad Sci USA 107(26):11865–11870. https://doi.org/10.1073/pnas.1005766107
Chen Z, Zou Y (2017) A multiscale model for heterogeneous tumor spheroid in vitro. Math Biosci Eng 15(2):361–392. https://doi.org/10.3934/mbe.2018016
Chu YS, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, Dufour S (2004) Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J Cell Biol 167(6):1183–1194. https://doi.org/10.1083/jcb.200403043
Cooper G (2007) The eukaryotic cell cycle. In: Cooper GM, Hausman RE (eds) The cell: a molecular approach. ASM Press, Washington, DC
Couzin ID, Krause J, Franks NR, Levin SA (2005) Effective leadership and decision-making in animal groups on the move. Nature 433(7025):513–516. https://doi.org/10.1038/nature03236
Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK (2008) AFM-based analysis of human metastatic cancer cells. Nanotechnology 19(38):384003. https://doi.org/10.1088/0957-4484/19/38/384003
Dickinson RB, Tranquillo RT (1993) Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE J 39(12):1995–2010. https://doi.org/10.1002/aic.690391210
DiMilla P, Barbee K, Lauffenburger D (1991) Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J 60(1):15–37. https://doi.org/10.1016/S0006-3495(91)82027-6
Drasdo D, Hoehme S (2012) Modeling the impact of granular embedding media, and pulling versus pushing cells on growing cell clones. New J Phys 14(5):055025. https://doi.org/10.1088/1367-2630/14/5/055025
Du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Siberzan P, Ladoux B (2005) Force mapping in epithelial cell migration. Proc Natl Acad Sci 102(7):2390–2395
DuChez BJ, Doyle AD, Dimitriadis EK, Yamada KM (2019) Durotaxis by human cancer cells. Biophys J 116(4):670–683. https://doi.org/10.1016/j.bpj.2019.01.009
Erdmann T, Schwarz US (2006) Bistability of cell-matrix adhesions resulting from nonlinear receptor-ligand dynamics. Biophys J 91(6):L60–L62
Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z (2008) Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell 14(4):570–581. https://doi.org/10.1016/j.devcel.2008.03.003
Fraley SI, Wu PH, He L, Feng Y, Krisnamurthy R, Longmore GD, Wirtz D (2015) Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions. Sci Rep 5(January):14580. https://doi.org/10.1038/srep14580
Frascoli F, Hughes BD, Zaman MH, Landman KA (2013) A computational model for collective cellular motion in three dimensions: general framework and case study for cell pair dynamics. PloS ONE 8(3):e59249. https://doi.org/10.1371/journal.pone.0059249
Friedl P, Mayor R (2017) Tuning collective cell migration by cell–cell junction regulation. Cold Spring Harb Perspect Biol 9(4):1146–1159. https://doi.org/10.1101/cshperspect.a029199
Friedl P, Noble PB, Walton PA, Laird DW, Chauvin PJ, Tabah RJ, Black M, Zanker KS (1995) Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res 55(20):4557–4560
Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E (2007) Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 9(12):1392–400. https://doi.org/10.1038/ncb1658
Gallant ND, Michael KE, García AJ (2005) Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol Biol Cell 16(9):4329–40. https://doi.org/10.1091/mbc.e05-02-0170
Garcia S, Hannezo E, Elgeti J, Joanny JF, Silberzan P, Gov NS (2015) Physics of active jamming during collective cellular motion in a monolayer. Proc Natl Acad Sci USA 112(50):15314–15319. https://doi.org/10.1073/pnas.1510973112
Gaudet C, Marganski WA, Kim S, Brown CT, Gunderia V, Dembo M, Wong JY (2003) Influence of type I collagen surface density on fibroblast spreading, motility, and contractility. Biophys J 85(5):3329
Gillitzer R, Goebeler M (2001) Chemokines in cutaneous wound healing. J Leukoc Biol 69(4):513–21
Goodenough AE, Little N, Carpenter WS, Hart AG (2017) Birds of a feather flock together: insights into starling murmuration behaviour revealed using citizen science. PLoS ONE 12(6):1–18. https://doi.org/10.1371/journal.pone.0179277
Harjanto D, Zaman MH (2013) Modeling extracellular matrix reorganization in 3d environments. PloS ONE 8(1):e52509
Ilina O, Bakker GJ, Vasaturo A, Hofmann RM, Friedl P (2011) Two-photon laser-generated microtracks in 3d collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Phys Biol 8(1):015010. https://doi.org/10.1088/1478-3975/8/1/015010
Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, Schulte-Merker S, Gerhardt H (2010) Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol 12(10):943–953. https://doi.org/10.1038/ncb2103
Kato T, Enomoto A, Watanabe T, Haga H, Ishida S, Kondo Y, Furukawa K, Urano T, Mii S, Weng L, Ishida-Takagishi M, Asai M, Asai N, Kaibuchi K, Murakumo Y, Takahashi M (2014) TRIM27/MRTF-B-dependent integrin \(\beta\)1 expression defines leading cells in cancer cell collectives. Cell Rep 7(4):1156–1167. https://doi.org/10.1016/j.celrep.2014.03.068
Kim MC, Whisler J, Silberberg YR, Kamm RD, Asada HH (2015) Cell invasion dynamics into a three dimensional extracellular matrix fibre network. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1004535
Knutsdottir H, Condeelis JS, Palsson E (2016) 3-D individual cell based computational modeling of tumor cell-macrophage paracrine signaling mediated by EGF and CSF-1 gradients. Integr Biol Quant Biosci Nano Macro 8(1):104–19. https://doi.org/10.1039/c5ib00201j
Lambert AW, Pattabiraman DR, Weinberg RA (2017) Emerging biological principles of metastasis. Cell 168(4):670–691. https://doi.org/10.1016/j.cell.2016.11.037
Lange JR, Fabry B (2013) Cell and tissue mechanics in cell migration. Exp Cell Res 319(16):2418–23. https://doi.org/10.1016/j.yexcr.2013.04.023
Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM (2009) Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139(5):891–906. https://doi.org/10.1016/j.cell.2009.10.027
Li F, Redick SD, Erickson HP, Moy VT (2003) Force measurements of the \(\alpha\)5\(\beta\)1 integrin–fibronectin interaction. Biophy J 84(2):1252–1262
Lo CM, Wang HB, Dembo M, Yl Wang (2000) Cell movement is guided by the rigidity of the substrate. Biophys J 79(1):144–152. https://doi.org/10.1016/S0006-3495(00)76279-5
Lusche DF, Wessels D, Soll DR (2009) The effects of extracellular calcium on motility, pseudopod and uropod formation, chemotaxis, and the cortical localization of myosin II in Dictyostelium discoideum. Cell Motil Cytoskelet 66(8):567–587. https://doi.org/10.1002/cm.20367
Mallon PSFNE (2001) Individual and collective decision-making during nest site selection by the ant Leptothorax albipennis. Behav Ecol Sociobiol 50(4):352–359. https://doi.org/10.1007/s002650100377
Mansury Y, Kimura M, Lobo J, Deisboeck TS (2002) Emerging patterns in tumor systems: simulating the dynamics of multicellular clusters with an agent-based spatial agglomeration model. J Theor Biol 219(3):343–370. https://doi.org/10.1006/jtbi.2002.3131
Morris BA, Burkel B, Ponik SM, Fan J, Condeelis JS, Aguirre-Ghiso JA, Castracane J, Denu JM, Keely PJ (2016) Collagen matrix density drives the metabolic shift in breast cancer cells. EBioMedicine 13:146–156. https://doi.org/10.1016/j.ebiom.2016.10.012
Mousavi SJ, Doweidar MH, Doblaré M (2014) Computational modelling and analysis of mechanical conditions on cell locomotion and cell–cell interaction. Comput Methods Biomech Biomed Eng 17(6):678–93. https://doi.org/10.1080/10255842.2012.710841
Munjal A, Lecuit T (2014) Actomyosin networks and tissue morphogenesis. Development 141(9):1789–93. https://doi.org/10.1242/dev.091645
Nasrollahi S, Pathak A (2016) Topographic confinement of epithelial clusters induces epithelial-to-mesenchymal transition in compliant matrices. Sci Rep 6(January):18831. https://doi.org/10.1038/srep18831
Palamidessi A, Malinverno C, Frittoli E, Corallino S, Barbieri E, Sigismund S, Beznoussenko GV, Martini E, Garre M, Ferrara I, Tripodo C, Ascione F, Cavalcanti-Adam EA, Li Q, Di Fiore PP, Parazzoli D, Giavazzi F, Cerbino R, Scita G (2019) Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma. Nat Mater 18(11):1252–1263. https://doi.org/10.1038/s41563-019-0425-1
Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF (1997) Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385(6616):537–540. https://doi.org/10.1038/385537a0
Palsson E (2001) A three-dimensional model of cell movement in multicellular systems. Future Gener Comput Syst 17(7):835–852. https://doi.org/10.1016/S0167-739X(00)00062-5
Palsson E (2008) A 3-D model used to explore how cell adhesion and stiffness affect cell sorting and movement in multicellular systems. J Theor Biol 254(1):1–13. https://doi.org/10.1016/j.jtbi.2008.05.004
Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE (2002) Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB J 16(10):1195–1204. https://doi.org/10.1096/fj.02-0038com
Petitjean L, Reffay M, Grasland-Mongrain E, Poujade M, Ladoux B, Buguin A, Silberzan P (2010) Velocity fields in a collectively migrating epithelium. Biophys J 98(9):1790–1800. https://doi.org/10.1016/j.bpj.2010.01.030
Plotnikov SV, Pasapera AM, Sabass B, Waterman CM (2012) Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151(7):1513–27. https://doi.org/10.1016/j.cell.2012.11.034
Reebs (2000) Can a minority of informed leaders determine the foraging movements of a fish shoal? Anim Behav 59(2):403–409. https://doi.org/10.1006/anbe.1999.1314
Robertson-Tessi M, Gillies RJ, Gatenby RA, Anderson ARA (2015) Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res 75(8):1567–1579. https://doi.org/10.1158/0008-5472.CAN-14-1428
Saez A, Buguin A, Silberzan P, Ladoux B (2005) Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J 89(6):L52–L54. https://doi.org/10.1529/biophysj.105.071217
Sepúlveda N, Petitjean L, Cochet O, Grasland-Mongrain E, Silberzan P, Hakim V (2013) Collective cell motion in an epithelial sheet can be quantitatively described by a stochastic interacting particle model. PLoS Computat Biol 9(3):e1002944. https://doi.org/10.1371/journal.pcbi.1002944
Sumpter DJT (2006) The principles of collective animal behaviour. Philos Trans R Soc Lond B Biol Sci 361(1465):5–22. https://doi.org/10.1098/rstb.2005.1733
Sun M, Bloom AB, Zaman MH (2015) Rapid quantification of 3d collagen fiber alignment and fiber intersection correlations with high sensitivity. PLoS ONE 10(7):1–17
Taubenberger A, Cisneros DA, Friedrichs J, Puech PH, Muller DJ, Franz CM (2007) Revealing early steps of \(\alpha\)2\(\beta\)1 integrin-mediated adhesion to collagen type I by using single-cell force spectroscopy. Mol Biol Cell 18(5):1634–1644
VanderVorst K, Dreyer CA, Konopelski SE, Lee H, Ho HYH, Carraway KL (2019) Wnt/PCP signaling contribution to carcinoma collective cell migration and metastasis. Cancer Res 79(8):1719–1729. https://doi.org/10.1158/0008-5472.CAN-18-2757
Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, Ladoux B (2012) Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad f Sci 109(32):12974–12979. https://doi.org/10.1073/pnas.1119313109
Velez DO, Tsui B, Goshia T, Chute CL, Han A, Carter H, Fraley SI (2017) 3d collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat Commun 8(1):1651. https://doi.org/10.1038/s41467-017-01556-7
Wang X, Enomoto A, Asai N, Kato T, Takahashi M (2016) Collective invasion of cancer: perspectives from pathology and development. Pathol Int 66(4):183–192. https://doi.org/10.1111/pin.12391
Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P (2007) Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. https://doi.org/10.1038/ncb1616
Wu PH, Giri A, Sun SX, Wirtz D (2014) Three-dimensional cell migration does not follow a random walk. Proc Natl Acad Sci USA 111(11):3949–54. https://doi.org/10.1073/pnas.1318967111
Yeoman BM, Katira P (2018) A stochastic algorithm for accurately predicting path persistence of cells migrating in 3d matrix environments. PloS ONE 13(11):e0207216
Zaman MH, Kamm RD, Matsudaira P, Lauffenburger Da (2005) Computational model for cell migration in three-dimensional matrices. Biophys J 89(2):1389–1397. https://doi.org/10.1529/biophysj.105.060723
Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P (2006) Migration of tumor cells in 3d matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci 103(29):10889–10894
Acknowledgements
This work was supported by Grants from the National Science Foundation (BMMB - 1905390, BMMB - 1763132 to P. K.) and the Army Research Office (W911NF-17-1-0413 to P. K.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest for any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10237_2020_1290_MOESM1_ESM.png
Figure S1. Mean Squared Displacement (MSD) Plots for Example Cells at 1.25x10-3 fibers/\(\mu m^3\) . A) MSD plot on a log–log scale for low alignment and low cell–cell adhesion. Fit equations are shown in the same color as their plot, with goodness of fit given by R. Solid blue line is defined leader and solid red line is undefined leader. Colored dashed lines are the fits for their matching color. Dashed black line is \(\langle R^2\rangle = \tau\). B) MSD plot on a log–log scale for high alignment and low cell–cell adhesion. C) MSD plot on a log–log scale for low alignment and high cell–cell adhesion. D) MSD plot on a log–log scale for high alignment and high cell–cell adhesion (png 294 KB)
10237_2020_1290_MOESM2_ESM.png
Figure S2. Mean Squared Displacement (MSD) Plots for Example Cells at 1.63x10-3 fibers/\(\mu m^3\) . A) MSD plot on a log–log scale for low alignment and low cell–cell adhesion. Fit equations are shown in the same color as their plot, with goodness of fit given by R. Solid blue line is defined leader and solid red line is undefined leader. Colored dashed lines are the fits for their matching color. Dashed black line is \(\langle R^2\rangle = \tau\). B) MSD plot on a log–log scale for high alignment and low cell–cell adhesion. C) MSD plot on a log–log scale for low alignment and high cell–cell adhesion. D) MSD plot on a log–log scale for high alignment and high cell–cell adhesion (png 293 KB)
10237_2020_1290_MOESM3_ESM.png
Figure S3. This figure shows MSD results for 5-cell cluster over increasing fiber density. Figure S1 (a), (c), and (e) shows MSD with no fiber alignment, and Figure S1 (b), (d), and (f) shows MSD for clusters with high fiber alignment. Figure S1 (a) and (b) shows MSD with clusters at low adhesion (25 nN), Figure S1 (c) and (d) shows MSD at medium adhesion (50 nN), and Figure S1 (e) and (f) shows MSD at high adhesion (100 nN) (png 761 KB)
10237_2020_1290_MOESM4_ESM.png
Figure S4. This figure shows MSD results for 10-cell cluster over increasing fiber density. Figure S2 (a), (c), and (e) shows MSD with no fiber alignment, and Figure S2 (b), (d), and (f) shows MSD for clusters with high fiber alignment. Figure S2 (a) and (b) shows MSD with clusters at low adhesion (25 nN), Figure S2 (c) and (d) shows MSD at medium adhesion (50 nN), and Figure S2 (e) and (f) shows MSD at high adhesion (100 nN) (png 732 KB)
10237_2020_1290_MOESM5_ESM.png
Figure S5. Cluster Size vs. Lifetime for Defined and Undefined Leading Clusters. A) Defined leader cluster lifetime as a function of cluster size. B) Undefined leader cluster as a function of cluster size. Clusters were simulated with low adhesion (25 nN) in unaligned matrices. Error bars are mean squared error (png 128 KB)
Video S1 shows the migration of a 10-cell cluster with a single defined leader over 18 hours. This sample was run with high adhesion, no fiber alignment, and an intermediate fiber density. The cell color at each time step corresponds to the phases accordingly: contraction (green), outgrowth (yellow), and retraction (red) (mp4 5948 KB)
Video S2 shows the migration of a 10-cell cluster with an undefined leader over 38 hours. This sample was run with high adhesion, no fiber alignment, and an intermediate fiber density. The color at each time step corresponds to the phases accordingly: contraction (green), outgrowth (yellow), and retraction (red) (mp4 1590 KB)
Rights and permissions
About this article
Cite this article
Collins, T.A., Yeoman, B.M. & Katira, P. To lead or to herd: optimal strategies for 3D collective migration of cell clusters. Biomech Model Mechanobiol 19, 1551–1564 (2020). https://doi.org/10.1007/s10237-020-01290-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10237-020-01290-y