Abstract
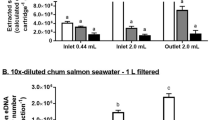
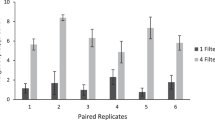
A simple environmental DNA (eDNA) filtration and extraction method that can be implemented on-site from water sampling to analysis was developed. In suspended glass fiber (SGF) method, 10 mg of the SGF was added to 1 L of environmental water to adsorb eDNA. The glass fiber was then collected using gravity filtration only, and eDNA was extracted using a simple reagent. eDNA was extracted from 85 samples of aquarium water, agricultural water, and river water using the SGF and standard methods. Under the laboratory condition, the SGF method was able to recover more DNA in the water containing lambda DNA than in the standard eDNA filtration and extraction method using adsorption columns. The invasive aquatic bivalve Limnoperna fortunei was used as a target species for detection using qPCR analysis or loop-mediated isothermal amplification (LAMP) analysis, which is less sensitive to DNA amplification inhibitors. The results showed that the positivity rate of the SGF-extracted DNA in qPCR and LAMP analysis was comparable or higher than that of the standard method extraction-qPCR analysis. In particular, as LAMP analysis is less time consuming than qPCR analysis and the presence or absence of DNA amplification can be determined visually, it is considered suitable as a rapid, on-site eDNA analysis method. The SGF method combined with the LAMP analysis has the potential to be applied to eDNA surveys of various species.





Similar content being viewed by others
References
Aichi Prefecture (2012) 50-year history of Aichi Prefectural waterworks and industrial waterworks (in Japanese). Aichi Prefectural Enterp Agency, Nagoya
Boulter N, Suarez FG, Schibeci S, Sunderland T, Tolhurst O, Hunter T, Hodge G, Handelsman D, Simanainen U, Hendriks E, Duggan K (2016) A simple, accurate and universal method for quantification of PCR. BMC Biotech 16:27. https://doi.org/10.1186/s12896-016-0256-y
Davis CN, Tyson F, Cutress D, Davies E, Jones DL, Brophy PM, Prescott A, Rose MT, Williams M, Williams HW, Jones RA (2020) Rapid detection of Galba truncatula in water sources on pasture-land using loop-mediated isothermal amplification for control of trematode infections. Parasites Vectors 13:496. https://doi.org/10.1186/s13071-020-04371-0
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425. https://doi.org/10.1098/rsbl.2008.0118
Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD, Schrenzel J (2011) Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 62:41–48
Fukumoto S, Ushimaru A, Minamoto T (2015) A basin-scale application of environmental DNA assessment for rare endemic species and closely related exotic species in rivers: a case study of giant salamanders in Japan. J Appl Ecol 52:358–365. https://doi.org/10.1111/1365-2664.12392
Goto M, Honda E, Ogura A, Nomoto A, Hanaki K (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxynaphthol blue. Biotechniques 46:167–172
Hartman LJ, Coyne SR, Norwood DA (2005) Development of a novel internal positive control for Taqman® based assays. Mol Cell Probes 19(1):51–59
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, Jackson CA, Lance RF, Mangan AM, Monroe EM, Piaggio AJ, Stokdyk JP, Wilson CC, Richter CA (2019) Reporting the limits of detection and quantification for environmental DNA assays. Environ DNA 2:271–282. https://doi.org/10.1002/edn3.29
Matsuoka K, Nishi H (2010) Invasive alien species, Limnoperna fortunei discovered from the River Ure-gawa, Shinshiro City, Japan (in Japanese). Sci Rep Toyohashi Mus Nat Hist 20:1–4
Melzak KA, Sherwood CS, Turner RFB, Haynes CA (1996) Driving forces for DNA adsorption to silica in perchlorate solutions. J Colloid Interface Sci 181:635–644
Minamoto T, Miya M, Sado T, Seino S, Doi H, Kondoh M, Nakamura K, Takahara T, Yamamoto S, Yamanaka H, Araki H, Iwasaki W, Kasai A, Masuda R, Uchii K (2021) An illustrated manual for environmental DNA research: water sampling guidelines and experimental protocols. Environ DNA 3:8–13. https://doi.org/10.1002/edn3.121
Mori Y, Kitao M, Tomita N, Notomi T (2004) Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods 59:145–157. https://doi.org/10.1016/j.jbbm.2003.12.005
Moushomi R, Wilgar G, Carvalho G, Creer S, Seymour M (2019) Environmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular. Sci Rep 9:12500. https://doi.org/10.1038/s41598-019-48984-7
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63
Nzelu CO, Gomez EA, Caceres AG, Sakurai T, Martini-Robles L, Uezato H, Mimori T, Katakura K, Hashiguchi Y, Kato H (2014) Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop 132:1–6
R Core T (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org. Accessed 23 Sept 2020
Schrader C, Schielke A, Ellerbroek L, Johne R (2012) PCR inhibitors—occurrence, properties and removal. J Appl Microbiol 113:1014–1026. https://doi.org/10.1111/j.1365-2672.2012.05384-x
Shiragane A, Uchida A, Uchida S (2012) Population dynamics and spatial distribution of the invasive mussel, Limnoperna fortunei, from its discovery to the present in the Yahagi River, Japan (in Japanese). Limnol Tokai Region Jpn 54:43–52
Takeshita D, Terui S, Ikeda K, Mitsuzuka T, Osathanunkul M, Minamoto T (2020) Projection range of eDNA analysis in marshes: a suggestion from the Siberian salamander (Salamandrella keyserlingii) inhabiting the Kushiro marsh. Japan Peerj 8:e9764. https://doi.org/10.7717/peerj.9764
Tsuji S, Nakao R, Saito M, Minamoto T, Akamatsu Y (2021) Pre-centrifugation before DNA extraction mitigates extraction efficiency reduction of environmental DNA caused by the preservative solution (benzalkonium chloride) remaining in the filters. Limnology 23:9–16. https://doi.org/10.1007/s10201-021-00676-w
Turner CR, Barnes MA, Xu CCY, Jones SE, Jerde CL, Lodge DM (2014) Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol Evol 5(7):676–684. https://doi.org/10.1111/2041-210X.12206
Williams MR, Stedtfeld RD, Engle C, Salach P, Fakher U, Stedtfeld T, Dreelin E, Jan Stevenson R, Latimore J, Hashsham SA (2017) Isothermal amplification of environmental DNA (eDNA) for direct field-based monitoring and laboratory confirmation of Dreissena sp. PLoS ONE 12(10):e0186462. https://doi.org/10.1371/journal.pone.0186462
Xia Z, Zhan A, Gao Y, Zhang L, Haffner DG, MacIsaac HJ (2018) Early detection of a highly invasive bivalve based on environmental DNA (eDNA). Biol Invasions 20:437–447. https://doi.org/10.1007/s10530-017-1545-7
Yamanaka H, Motozawa H, Tsuji S, Miyazawa RC, Takahara T, Minamoto T (2016) On-site filtration of water samples for environmental DNA analysis to avoid DNA degradation during transportation. Ecol Res 31:963–967. https://doi.org/10.1007/s11284-016-1400-9
Acknowledgements
We would like to thank Toyota Land Improvement District, Japan Water Agency Aichi Canal General Management Office, and Japan Water Agency Toyogawa Canal Headquarters for their cooperation in collecting agricultural water. This work was supported by a grant from commissioned project study on “Development of management strategies for invasive alien species causing damage to agriculture” JPJ007966, Ministry of Agriculture, Forestry and Fisheries, Japan. The experiments in this study comply with current Japanese laws.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling Editor: Teruhiko Takahara.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suzuki, R., Kawamura, K. & Mizukami, Y. Simple extraction and analysis of environmental DNA using glass fibers in suspension form. Limnology 24, 25–36 (2023). https://doi.org/10.1007/s10201-022-00705-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10201-022-00705-2