Abstract
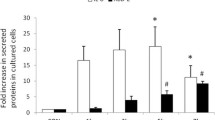
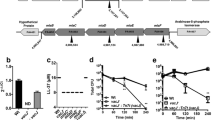
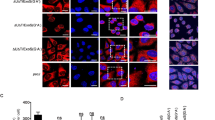
We investigated the influence of the type III effector, ExoS, on the host epithelial cell response to Pseudomonas aeruginosa infection, and we found that disruption of the exoS gene caused a significant increase in the amount of interleukin-8 (IL-8) in the culture medium of Caco-2 cells. We show that IL-8 was degraded in the culture medium following infection of the cells with the wild-type (PAO1), but not the exoS knock-out (the ΔexoS) strain. Purified ExoS protein itself did not degrade IL-8. We next show that IL-8 degradation by PAO1 was inhibited by the addition of serine protease inhibitors. These results strongly suggest that a bacterial serine protease that degrades IL-8 is expressed and secreted into the culture medium of Caco-2 cells infected with PAO1, and that the expression of this protein is repressed in cells infected with the ΔexoS strain. The PAO1 genome encodes 28 different protease genes, including two serine proteases: PA3535 and mucD. PA3535 and mucD gene knock-outs were constructed (ΔmucD and ΔPA3535), and ΔmucD but not ΔPA3535 showed reduced IL-8 degradation. To understand the significance of IL-8 degradation, we next evaluated neutrophil infiltration in lungs excised from mice intranasally infected with the P. aeruginosa strains. Increased neutrophil infiltration was observed in PAO1-infected mice, but not in ΔexoS- or ΔmucD-infected mice. Taken together, our results suggest that P. aeruginosa escapes from phagocytic killing due to IL-8 degradation following the secretion of the MucD serine protease, whose expression appears to be influenced by ExoS.









Similar content being viewed by others
References
Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J Clin Microbiol. 2003;41:3526–31.
Galán JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–8.
Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216.
Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34.
Raoust E, Balloy V, Garcia-Verdugo I, Touqui L, Ramphal R, Chignard M. Pseudomonas aeruginosa LPS or flagellin are sufficient to activate TLR-dependent signaling in murine alveolar macrophages and airway epithelial cells. PLoS One. 2009;4:e7259.
Cigana C, Curcurù L, Leone MR, Ieranò T, Lorè NI, Bianconi I, et al. Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One. 2009;4:e8439.
Delgado MA, Poschet JF, Deretic V. Nonclassical pathway of Pseudomonas aeruginosa DNA-induced interleukin-8 secretion in cystic fibrosis airway epithelial cells. Infect Immun. 2006;74:2975–84.
Pena J, Fu Z, Schwarzer C, Machen TE. Pseudomonas aeruginosa inhibition of flagellin-activated NF-kappaB and interleukin-8 by human airway epithelial cells. Infect Immun. 2009;77:2857–65.
Furuya N, Hirakata Y, Tomono K, Matsumoto T, Tateda K, Kaku M, et al. Mortality rates amongst mice with endogenous septicaemia caused by Pseudomonas aeruginosa isolates from various clinical sources. J Med Microbiol. 1993;39:141–6.
Hirakata Y, Srikumar R, Poole K, Gotoh N, Suematsu T, Kohno S, et al. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp Med. 2002;196:109–18.
Matsumoto T, Tateda K, Miyazaki S, Furuya N, Ohno A, Ishii Y, et al. Adverse effects of tumour necrosis factor in cyclophosphamide-treated mice subjected to gut-derived Pseudomonas aeruginosa sepsis. Cytokine. 1997;9:763–9.
Matsumoto T, Tateda K, Miyazaki S, Furuya N, Ohno A, Ishii Y, et al. Effect of interleukin-10 on gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob Agents Chemother. 1998;42:2853–7.
Goehring UM, Schmidt G, Pederson KJ, Aktories K, Barbieri JT. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J Biol Chem. 1999;274:36369–72.
Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7:654–65.
Henriksson ML, Francis MS, Peden A, Aili M, Stefansson K, Palmer R, et al. A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur J Biochem. 2002;269:4921–9.
Würtele M, Renault L, Barbieri JT, Wittinghofer A, Wolf E. Structure of the ExoS GTPase activating domain. FEBS Lett. 2001;491:26–9.
Coburn J, Gill DM. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–62.
Coburn J, Wyatt RT, Iglewski BH, Gill DM. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–8.
Henriksson ML, Sundin C, Jansson AL, Forsberg A, Palmer RH, Hallberg B. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem J. 2002;367:617–28.
Okuda J, Hayashi N, Okamoto M, Sawada S, Minagawa S, Yano Y, et al. Translocation of Pseudomonas aeruginosa from the intestinal tract is mediated by the binding of ExoS to an Na, K-ATPase regulator, FXYD3. Infect Immun. 2010;78:4511–22.
Sutterawala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–45.
Okuda J, Toyotome T, Kataoka N, Ohno M, Abe H, Shimura Y, et al. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem Biophys Res Commun. 2005;333:531–9.
Ferguson MW, Maxwell JA, Vincent TS, da Silva J, Olson JC. Comparison of the exoS gene and protein expression in soil and clinical isolates of Pseudomonas aeruginosa. Infect Immun. 2001;69:2198–210.
Edwards RJ, Taylor GW, Ferguson M, Murray S, Rendell N, Wrigley A, et al. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J Infect Dis. 2005;192:783–90.
Hidalgo-Grass C, Mishalian I, Dan-Goor M, Belotserkovsky I, Eran Y, Nizet V, et al. A streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 2006;25:4628–37.
Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, et al. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe. 2008;4:170–8.
Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–79.
Wood LF, Ohman DE. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J Bacteriol. 2006;188:3134–7.
Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–23.
Hobert ME, Sands KA, Mrsny RJ, Madara JL. Cdc42 and Rac1 regulate late events in Salmonella typhimurium-induced interleukin-8 secretion from polarized epithelial cells. J Biol Chem. 2002;277:51025–32.
Acknowledgments
This work was supported in part by a Kyoto Pharmaceutical University Fund for the Promotion of Scientific Research.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Okuda, J., Hayashi, N., Tanabe, S. et al. Degradation of interleukin 8 by the serine protease MucD of Pseudomonas aeruginosa . J Infect Chemother 17, 782–792 (2011). https://doi.org/10.1007/s10156-011-0257-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10156-011-0257-7