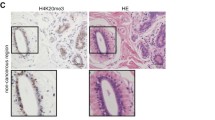
Abstract
Background
Results of previous studies about the prognostic roles of histone H4 lysine 16 acetylation (H4K16ac) and histone H4 lysine 20 trimethylation (H4K20me3) in breast cancer were inconsistent. Cellular experiments revealed the interplays between H4K16ac and H4K20me3, but no population study explored the interaction between them on the prognosis.
Methods
H4K16ac and H4K20me3 levels in tumors were evaluated by immunohistochemistry for 958 breast cancer patients. Hazard ratios for overall survival (OS) and progression-free survival (PFS) were estimated using Cox regression models. Interaction was assessed on multiplicative scale. Concordance index (C-index) was calculated to verify the predictive performance.
Results
The prognostic roles of the low level of H4K16ac or H4K20me3 were significant only in patients with the low level of another marker and their interactions were significant. Moreover, compared with joint high levels of both them, only the combined low levels of both them was associated with a poor prognosis but not the low level of single one. The C-index of the clinicopathological model combined the joint expression of H4K16ac and H4K20me3 [0.739 for OS; 0.672 for PFS] was significantly larger than that of the single clinicopathological model [0.699 for OS, P < 0.001; 0.642 for PFS, P = 0.003] or the model combined with the single H4K16ac [0.712 for OS, P < 0.001; 0.646 for PFS, P < 0.001] or H4K20me3 [0.724 for OS, P = 0.031; 0.662 for PFS, P = 0.006].
Conclusions
There was an interaction between H4K16ac and H4K20me3 on the prognosis of breast cancer and the combination of them was a superior prognostic marker compared to the single one.




Similar content being viewed by others
References
Campbell LL, Polyak K (2007) Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 6:2332–2338. https://doi.org/10.4161/cc.6.19.4914
De Abreu FB, Schwartz GN, Wells WA et al (2014) Personalized therapy for breast cancer. Clin Genet 86:62–67. https://doi.org/10.1111/cge.12381
Kunc M, Peksa R, Cserni G et al (2022) High expression of progesterone receptor may be an adverse prognostic factor in oestrogen receptor-negative/progesterone receptor-positive breast cancer: results of comprehensive re-evaluation of multi-institutional case series. Pathology 54:269–278. https://doi.org/10.1016/j.pathol.2021.10.003
Han Y, Wu Y, Xu H et al (2022) The impact of hormone receptor on the clinical outcomes of HER2-positive breast cancer: a population-based study. Int J Clin Oncol 27:707–716. https://doi.org/10.1007/s10147-022-02115-x
Siegel RL, Miller KD, Fuchs HE et al (2021) Cancer Statistics, 2021. CA Cancer J Clin 71:7–33. https://doi.org/10.3322/caac.21654
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Piekarz RL, Bates SE (2009) Epigenetic modifiers: basic understanding and clinical development. Clin Cancer Res 15:3918–3926. https://doi.org/10.1158/1078-0432.CCR-08-2788
Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31:27–36. https://doi.org/10.1093/carcin/bgp220
Fraga MF, Ballestar E, Villar-Garea A et al (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391–400. https://doi.org/10.1038/ng1531
Kapoor-Vazirani P, Kagey JD, Powell DR et al (2008) Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res 68:6810–6821. https://doi.org/10.1158/0008-5472.CAN-08-0141
Zhao LJ, Loewenstein PM, Green M (2016) The adenoviral E1A N-terminal domain represses MYC transcription in human cancer cells by targeting both p300 and TRRAP and inhibiting MYC promoter acetylation of H3K18 and H4K16. Genes Cancer 7:98–109. https://doi.org/10.18632/genesandcancer.99
Shinchi Y, Hieda M, Nishioka Y et al (2015) SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp Cell Res 334:90–99. https://doi.org/10.1016/j.yexcr.2015.03.010
Kapoor-Vazirani P, Kagey JD, Vertino PM (2011) SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 31:1594–1609. https://doi.org/10.1128/MCB.00524-10
Elsheikh SE, Green AR, Rakha EA et al (2009) Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res 69:3802–3809. https://doi.org/10.1158/0008-5472.CAN-08-3907
Yokoyama Y, Matsumoto A, Hieda M et al (2014) Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res 16:R66. https://doi.org/10.1186/bcr3681
Wolff AC, Hammond ME, Schwartz JN et al (2007) American society of clinical oncology/college of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25:118–145. https://doi.org/10.1200/JCO.2006.09.2775
Chen QX, Yang YZ, Liang ZZ et al (2021) Time-varying effects of FOXA1 on breast cancer prognosis. Breast Cancer Res Treat 187:867–875. https://doi.org/10.1007/s10549-021-06125-7
See SHC, Smith SH, Finkelman BS et al (2023) The role of PRAME and NY-ESO-1 as potential therapeutic and prognostic biomarkers in triple-negative breast carcinomas. Pathol Res Pract 241:154299. https://doi.org/10.1016/j.prp.2022.154299
Li K, Zhao G, Yuan H et al (2022) Upregulated expression of DDX5 predicts recurrence and poor prognosis in breast cancer. Pathol Res Pract 229:153736. https://doi.org/10.1016/j.prp.2021.153736
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10:7252–7259. https://doi.org/10.1158/1078-0432.CCR-04-0713
Chen D, Liu Z, Liu W et al (2021) Predicting postoperative peritoneal metastasis in gastric cancer with serosal invasion using a collagen nomogram. Nat Commun 12:179. https://doi.org/10.1038/s41467-020-20429-0
Chen L, Zeng H, Du Z et al (2020) Nomogram based on pre-treatment inflammatory biomarkers predicting survival in patients with head and neck soft tissue sarcoma. Cancer Biomark 29:151–161. https://doi.org/10.3233/CBM-201739
Wang Y, Li J, Xia Y et al (2013) Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol 31:1188–1195. https://doi.org/10.1200/JCO.2012.41.5984
Huitzil-Melendez FD, Capanu M, O’Reilly EM et al (2010) Advanced hepatocellular carcinoma: which staging systems best predict prognosis? J Clin Oncol 28:2889–2895. https://doi.org/10.1200/JCO.2009.25.9895
Conway KE, McConnell BB, Bowring CE et al (2000) TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res 60:6236–6242
Parsons MJ, Vertino PM (2006) Dual role of TMS1/ASC in death receptor signaling. Oncogene 25:6948–6958. https://doi.org/10.1038/sj.onc.1209684
Qian X, Li G, Vass WC et al (2009) The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell 16:246–258. https://doi.org/10.1016/j.ccr.2009.07.031
Pasculli B, Barbano R, Parrella P (2018) Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin Cancer Biol 51:22–35. https://doi.org/10.1016/j.semcancer.2018.01.007
Damaskos C, Garmpis N, Valsami S et al (2017) Histone deacetylase inhibitors: an attractive therapeutic strategy against breast cancer. Anticancer Res 37:35–46. https://doi.org/10.21873/anticanres.11286
Connolly RM, Zhao F, Miller KD et al (2021) E2112: randomized phase III trial of endocrine therapy plus entinostat or placebo in hormone receptor-positive advanced breast cancer. A trial of the ECOG-ACRIN cancer research group. J Clin Oncol 39:3171–3181. https://doi.org/10.1200/JCO.21.00944
Arrowsmith CH, Bountra C, Fish PV et al (2012) Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 11:384–400. https://doi.org/10.1038/nrd3674
Connolly R, Stearns V (2012) Epigenetics as a therapeutic target in breast cancer. J Mammary Gland Biol Neoplasia 17:191–204. https://doi.org/10.1007/s10911-012-9263-3
Saji S, Kimura-Tsuchiya R (2015) Combination of molecular-targeted drugs with endocrine therapy for hormone-resistant breast cancer. Int J Clin Oncol 20:268–272. https://doi.org/10.1007/s10147-015-0799-2
Collins HM, Abdelghany MK, Messmer M et al (2013) Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer 13:37. https://doi.org/10.1186/1471-2407-13-37
Balasubramanyam K, Altaf M, Varier RA et al (2004) Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem 279:33716–33726. https://doi.org/10.1074/jbc.M402839200
Aggarwal V, Tuli HS, Kaur J et al (2020) Garcinol exhibits anti-neoplastic effects by targeting diverse oncogenic factors in tumor cells. Biomedicines 8:103. https://doi.org/10.3390/biomedicines8050103
Sanchez-Munoz A, Perez-Ruiz E, Jurado JM et al (2011) Outcome of small invasive breast cancer with no axillary lymph node involvement. Breast J 17:32–38. https://doi.org/10.1111/j.1524-4741.2010.01026.x
Xu H, Jin F, Zhang XJ et al (2020) Adherence status to adjuvant endocrine therapy in Chinese women with early breast cancer and its influencing factors: a cross-sectional survey. Cancer Med 9:3703–3713. https://doi.org/10.1002/cam4.3017
Acknowledgements
We sincerely thank the patients who participated in this study, the staff who conducted the baseline and the follow-up data collection, and the medical staff in the breast departments of the Third Affiliated Hospital, and the Cancer Center of Sun Yat-Sen University. We also sincerely thank the funding of National Natural Science Foundation of China (81973115) and Science and Technology Planning Project of Guangdong Province, China (2019B030316002).
Author information
Authors and Affiliations
Contributions
BW, MZ and ZFR designed and directed the study, wrote and/or revised the manuscript. YZY constructed the TMAs and contributed to the IHC. YXR, ZJW, XFZ, JXG, and LYT contributed to digital imaging of IHC-stained sections and the assessment of immunohistochemical expression. BW, MZ and XLG contributed to clinical data collection and curation. BW and MZ participated in the statistical analysis plan and interpretation of results. ZFR provided administrative support and supervision for the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1: Figure S1.
Flow chart of the study cohort. Note: there were three people who missed the data of both H4K16ac and H4K20me3 level. H4K16ac, Histone H4 lysine 16 acetylation; H4K20me3, Histone H4 lysine 20 trimethylation. Figure S2. Restricted cubic splines of H4K16ac and H4K20me3 with breast cancer OS (a for H4K16ac, c for H4K20me3) and PFS (b for H4K16ac, d for H4K20me3). H4K16ac, Histone H4 lysine 16 acetylation; H4K20me3, Histone H4 lysine 20 trimethylation; OS, overall survival; PFS, progression-free survival. Figure S3. Kaplan-Meier survival curves of H4K16ac and H4K20me3 with breast cancer OS (a for H4K16ac, c for H4K20me3) and PFS (b for H4K16ac, d for H4K20me3). H4K16ac, Histone H4 lysine 16 acetylation; H4K20me3, Histone H4 lysine 20 trimethylation; OS, overall survival; PFS, progression-free survival. Table S1. Univariate association between the demographic and clinicopathological characteristics and the outcomes. Table S2. Univariate and multivariate association between H4K16ac and H4K20me3 levels in tumor tissues and the outcomes.
About this article
Cite this article
Wang, B., Zhou, M., Gan, Xl. et al. Combined low levels of H4K16ac and H4K20me3 predicts poor prognosis in breast cancer. Int J Clin Oncol 28, 1147–1157 (2023). https://doi.org/10.1007/s10147-023-02378-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-023-02378-y