Abstract
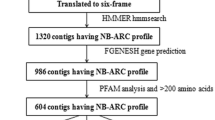
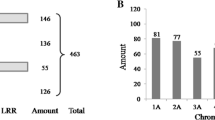
Banana is an important food crop that is susceptible to a wide range of pests and diseases that can reduce yield and quality. The primary objective of banana breeding programs is to increase disease resistance, which requires the identification of resistance (R) genes. Despite the fact that resistant sources have been identified in bananas, the genes, particularly the nucleotide-binding site (NBS) family, which play an important role in protecting plants against pathogens, have received little attention. As a result, this study included a thorough examination of the NBS disease resistance gene family’s classification, phylogenetic analysis, genome organization, evolution, cis-elements, differential expression, regulation by microRNAs, and protein–protein interaction. A total of 116 and 43 putative NBS genes from M. acuminata and M. balbisiana, respectively, were identified and characterized, and were classified into seven sub-families. Structural analysis of NBS genes revealed the presence of signal peptides, their sub-cellular localization, molecular weight and pI. Eight commonly conserved motifs were found, and NBS genes were unevenly distributed across multiple chromosomes, with the majority of NBS genes being located in chr3 and chr1 of the A and B genomes, respectively. Tandem duplication occurrences have helped bananas’ NBS genes spread throughout evolution. Transcriptome analysis of NBS genes revealed significant differences in expression between resistant and susceptible cultivars of fusarium wilt, eumusae leaf spot, root lesion nematode, and drought, implying that they can be used as candidate resistant genes. Ninety miRNAs were discovered to have targets in 104 NBS genes from the A genome, providing important insights into NBS gene expression regulation. Overall, this study offers a valuable genomic resource and understanding of the function and evolution of NBS genes in relation to rapidly evolving pathogens, as well as providing breeders with selection targets for fast-tracking breeding of banana varieties with more durable resistance to pathogens.





Similar content being viewed by others
Data availability
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request. All other data generated or analyzed during this study are included in this article supplementary information files.
Abbreviations
- NBS:
-
Nucleotide-binding site
- PTI:
-
Pathogen-triggered immunity
- ETI:
-
Effector-triggered immunity
- SAR:
-
Systemic acquired resistance
- PAMPs:
-
Pathogen-associated molecular patterns
- PRRs:
-
Pattern recognition receptors
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Ameline-Torregrosa C, Wang B, O’Bleness MS, Deshpande S, Zhu H, Roe BA, Young ND, Cannon SB (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:21–25
Andersen EJ, Ali S, Reese RN, Yen Y, Neupane S, Nepal MP (2016) Diversity and evolution of disease resistance genes in barley (Hordeum vulgare L.). Evol Bio Inform 12:99–108
Anuradha C, Chandrasekar A, Backiyarani S, Uma S (2022) MusaRgeneDB: An online comprehensive database for disease resistance genes in Musa spp 3. Biotech 12:1–12
Anuradha C, Chandrasekar A, Backiyarani S, Thangavelu R, Giribabu P, Uma S (2022) Genome-wide analysis of pathogenesis-related protein 1 (PR-1) gene family from Musa spp and its role in defense response during stresses. Gene 821:146334
Arisha MH, Ahmad MQ, Tang W, Liu Y, Yan H, Kou M, Wang X, Zhang Y, Li Q (2020) RNA-sequencing analysis revealed genes associated drought stress responses of different durations in hexaploid sweet potato. Sci Rep 10:12573
Ashburner M, Ball CA, Blake JA et al (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29
Ayliffe MA, Lagudah ES (2004) Molecular genetics of disease resistance in cereals. Ann Bot 94(6):765–773
Backiyarani S, Uma S, Arunkumar G, Saraswathi MS, Sundararaju P (2013) Cloning and characterization of NBS-LRR resistance gene analogues of Musa spp. and their expression profiling studies against Pratylenchus coffeae. Afr J Biotechnol 12:4256–4268
Bai J, Pennill LA, Ning J et al (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12:1871–1884
Baumgarten A, Cannon S, Spangler R, May G (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165:309–319
Buchel AS, Brederode FT, Bol JF, Linthorst HJ (1999) Mutation of GT-1 binding sites in the PR-1A promoter influences the level of inducible gene expression in vivo. Plant Mol Biol 40:387–396
Cannon SB, Mitra A, Baumgarten A et al (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10. https://doi.org/10.1186/1471-2229-4-10
Chang W, Li H, Chen H, Qiao F, Zeng H (2020) NBS-LRR gene family in banana (Musa acuminata): genome-wide identification and responses to Fusarium oxysporum f. sp. cubense race 1 and tropical race 4. Eur J Plant Pathol 157:549–563
Chen Z, Zhao W, Zhu X et al (2018) Identification and characterization of rice blast resistance gene Pid4 by a combination of transcriptomic profiling and genome analysis. J Genet Genomics 45(12):663–672
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J Cell Mol Biol 38(5):810–822
Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14(10):521–529
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Cortaga CQ, Latina RA, Habunal RR, Lantican DV (2022) Identification and characterization of genome-wide resistance gene analogs (RGAs) of durian (Durio zibethinus L.). J Genet Eng Biotechnol 20:29
Cristina MS, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649
Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66:487–511
Dale J, James A, Paul JY et al (2017) Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat Commun 8(1):1496
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Davey MW, Van den Bergh I, Markham R, Swennen R, Keulemans J (2009) Genetic variability in Musa fruit provitaminA carotenoids, lutein and mineral micronutrient contents. Food Chem 115(3):806–813
Davey M, Gudimella R, Harikrishna JA, Sin LW, Khalid N, Keulemans J (2013) A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genomics 14:683
D’Hont A, Denoeud F, Aury JM et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488(7410):213–217
Die JV, Román B, Qi X, Rowland LJ (2018) Genome-scale examination of NBS-encoding genes in blueberry. Sci Rep 8(1):3429
Dodds PN, Lawrence GJ, Ellis JG (2001) Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13:163–178
Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51:21–37
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14(9):755–763
Eitas TK, Dangl JL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13(4):472–477
Emediato FL, Passos M, Teixeira CD, Pappas GJ, Miller RN (2013) Analysis of expression Of NBS-LRR resistance gene analogs in Musa acuminata during compatible and incompatible interactions with Mycosphaerella musicola. Acta Hort 986:255–258
Eulgem T, Rushton P, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Faigón-Soverna A, Harmon FG, Storani L et al (2006) A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell 18:2919–2928
Friedman AR, Baker BJ (2007) The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev 17:493–499
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64:839–863
Fu Y, Zhang Y, Mason AS, Lin B, Zhang D, Yu H, Fu D (2019) NBS-encoding genes in Brassica napus evolved rapidly after allopolyploidization and co-localize with known disease resistance loci. Front Plant Sci 10:26
Goyal N, Bhatia G, Sharma S, Garewal N, Upadhyay A, Upadhyay SK, Singh K (2020) Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 112(1):312–322
Gu L, Si W, Zhao L, Yang S, Zhang X (2015) Dynamic evolution of NBS–LRR genes in bread wheat and its progenitors. Mol Genet Genom 290:727–738
Guo Y, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157:757–769
Habachi-Houimli Y, Khalfallah Y, Mezghani-Khemakhem M, Makni H, Makni M, Bouktila D (2018) Genome-wide identification, characterization, and evolutionary analysis of NBS-encoding resistance genes in barley. 3Biotech 8:1–16
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Series 41:95–98
Holub EB (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2:516–527
Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z (2015) The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci 6:742
JiaY YY, Zhang Y, Yang S, Zhang X (2015) Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet 16:48
Jo B, Choi SS (2015) Introns: the functional benefits of introns in genomes. Genomics Inform 13(4):112–118
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. https://doi.org/10.1038/nature05286
Kang YJ, Kim K, Shim S, Yoon MY, Sun S, Kim MY, Van K, Lee S (2012) Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol 12:139–139
Kim Y, Chae S, Oh NI, Nguyen NH, Cheong J (2020) Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front Genet 11:576086
Koroban NV, Kudryavtseva AV, Krasnov GS et al (2016) The role of microRNA in abiotic stress response in plants. Mol Biol 50:337–343
Kourelis J, van der Hoorn RA (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Lan T, Yang Z, Yang X, Liu Y, Wang X, Zeng Q (2009) Extensive functional diversification of the populus glutathione s-transferase supergene family. The Plant Cell Online 21(12):3749–3766
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–8
Lescot M, Piffanelli P, Ciampi AY et al (2008) Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9:58
Lescot M, Ciampi AY, Ruiz M et al (2005) Fresh insights into the Musa genome and its comparison with rice. 2005.
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense on-line version contains web-only data. The Plant Cell Online 16:319–331
Li J, Ding J, Zhang WG, Zhang Y, Tang P, Chen J, Tian D, Yang S (2010a) Unique evolutionary pattern of numbers of gramineous NBS–LRR genes. Mol Genet Genom 283:427–438
Li X, Cheng Y, Ma W et al (2010b) Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst Evol 289(1–2):101–110
Li Y, Zhong Y, Huang K, Cheng Z (2016) Genomewide analysis of NBS-encoding genes in kiwi fruit (Actinidiachinensis). J Genet 95:997–1001
Lin X, Zhang Y, Kuang H, Chen J (2013) Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae. BMC Genom 14:335–335
Liu H, Dai J, Feng D, Liu B, Wang H, Wang J (2010) Characterization of a novel plantain Asr gene, MpAsr, that is regulated in response to infection of Fusarium oxysporum f. sp. cubense and abiotic stresses. J Integr Plant Biol 52(3):315–23
Liu F, Li H, Wu J, Wang B, Tian N, Liu J, Sun X, Wu H, Huang Y, Lü P, Cheng C (2021a) Genome-wide identification and expression pattern analysis of lipoxygenase gene family in banana. Sci Rep 11:9948
Liu Y, Li D, Yang N, Zhu X, Han K, Gu R, Bai J, Wang A, Zhang Y (2021b) Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in Chinese cabbage. Int J Mol Sci 22:4266. https://doi.org/10.3390/ijms22084266
Long M, VanKuren NW, Chen S, Vibranovski MD (2013) New gene evolution: little did we know. Annu Rev Genet 47:307–333
Lozano R, Hamblin MT, Prochnik S, Jannink J (2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16(2015):360
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155
Mace ES, Tai S, Innes DJ et al (2014) The plasticity of NBS resistance genes in sorghum is driven by multiple evolutionary processes. BMC Plant Biol 14:253
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32:77–92
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW et al (2003) Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15:809–834
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. CurrOpin Plant Biol 8:129–134
Miller RN, Bertioli DJ, Baurens FC, Santos CM, Alves PC, Martins NF, Togawa RC, Souza MT, Pappas GJ (2008) Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminate Colla: isolation, RFLP marker development, and physical mapping. BMC Plant Biol 8:15
Mondragon-Palomino M, Meyers BC, Michelmore RW, Gaut BS (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12:1305–1315
Mun J, Yu H, Park S, Park B (2009) Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genom 282:617–631
Muthusamy M, Uma S, Backiyarani S, Saraswathi MS, Chandrasekar A (2016) Transcriptomic changes of drought-tolerant and sensitive banana cultivars exposed to drought stress. Front Plant Sci 7:1609
Nansamba M, Sibiya J, Tumuhimbise R, Karamura D, Kubiriba J, Karamura E (2020) Breeding banana (Musa spp) for drought tolerance a review. Plant Breed 139:685–696
Ouyang S, Park G, Atamian HS, Han CS, Stajich JE, Kaloshian I, Borkovich KA (2014) MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog 10:e1004464
Pan Q, Wendel JF, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J MolEvol 50:203–213
Park JH, Shin C (2015) The role of plant small RNAs in NB-LRR regulation. Brief Funct Genom 14:268–274
Passos M, de Cruz VO, Emediato FL et al (2013) Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycosphaerella musicola: gene assembly, annotation and marker development. BMC Genomics 14:78
Peraza-Echeverria S, Dale JL, Harding RM, Smith MK, Collet C (2008) Characterization of disease resistance gene candidates of the nucleotide binding site (NBS) type from banana and correlation of a transcriptional polymorphism with resistance to Fusarium oxysporum f. sp. cubense race 4. Mol Breed 22(4):565–579
Peraza-Echeverria S, Dale JL, Harding RM, Collet C (2009) Molecular cloning and in silico analysis of potential Fusarium resistance genes in banana. Mol Breed 23:431–443
Porter BW, Paidi MD, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics 281:609–626
Qian L, Zhou G, Sun X, Lei Z, Zhang Y, Xue J, Hang Y (2017) Distinct patterns of gene gain and loss diverse evolutionary modes of NBS-encoding genes in three solanaceae crop species. G3-Genes Genom Genet 7(5):1577–1585
Ravi I, Uma S, Vaganan MM, Mustaffa MM (2013) Phenotyping bananas for drought resistance. Front Physiol 4:9
Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19(1):76–84
Roy SW, Gilbert W (2005) Rates of intron loss and gain: implications for early eukaryotic evolution. Proc Natl Acad Sci USA 102(16):5773–5778
Rushton P, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15:5690–5700
Rushton P, Reinstädler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. The Plant Cell Online 14:749–762
Santaella M, Suárez E, López CE, González C, Mosquera GM, Restrepo S, Tohme J, Badillo A, Verdier V (2004) Identification of genes in cassava that are differentially expressed during infection with Xanthomonas axonopodis pv. manihotis. Mol Plant Pathol 5(6):549–58
Santamaria M, Thomson CJ, Read ND, Loake GJ (2001) The promoter of a basic PR1-like gene, AtPRB1, from Arabidopsis establishes an organ-specific expression pattern and responsiveness to ethylene and methyl jasmonate. Plant Mol Biol 47:641–652
Saravanakumar AS, Uma S, Thangavelu R, Backiyrani S, Saraswathi MS, Sriram V (2016) Preliminary analysis on the transcripts involved in resistance responses to eumusae leaf spot disease of banana caused by Mycosphaerellaeumusae, a recent add-on of the sigatoka disease complex. Turk J Botany 40(5):461–471
Schutter BD, Speijer PR, Dochez C, Tenkouano A, Waele DD (2001) Evaluating host plant reaction of Musa germplasm to Radopholussimilis by inoculation of single primary roots. Nematropica 31(2):295–300
Shao Z, Zhang Y, Hang Y, Xue J, Zhou G, Wu P, Wu X, Wu X, Wang Q, Wang B, Chen J (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol 166(1):217–234
Shao Z, Xue J, Wu P, Zhang Y, Wu Y, Hang Y, Wang B, Chen J (2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170:2095–2109
Shao Z, Xue J, Wang Q, Wang B, Chen J (2019) Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci 24(1):9–12
Shiu S, Karłowski WM, Pan R, Tzeng Y, Mayer KF, Li W (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16(5):1220–1234
Sutanto A, Sukma D, Hermanto C, Sudarsono SA (2014) Isolation and characterization of resistance gene analogue (RGA) from Fusarium resistant banana cultivars. Emir J Food Agric 26:508–518
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613
Tan S, Wu S (2012) Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp Funct Genomics 2012:418208
Thangavelu R, Saraswathi MS, Uma S, Loganathan M, Backiyarani S, Durai P, Raj E, Marimuthu N, Kannan G, Swennen R (2021) Identification of sources resistant to a virulent Fusarium wilt strain (VCG 0124) infecting Cavendish bananas. Sci Rep 11:3183
Timm ES, Pardo LH, Coello RP, Navarrete TC, Villegas ON, Ordoñez EG (2016) Identification of differentially-expressed genes in response to Mycosphaerella fijiensisin the resistant Musa accession ‘Calcutta-4’ using suppression subtractive hybridization. PLoS ONE 11(8):e0160083
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur J Biochem 222(1):9–19
Vanhove AC, Vermaelen W, Panis B, Swennen R, Carpentier SC (2012) Screening the banana biodiversity for drought tolerance: can an in vitro growth model and proteomics be used as a tool to discover tolerant varieties and understand homeostasis. Front Plant Sci 3:176
Wan H, Yuan W, Bo K, Shen J, Pang X, Chen J (2013) Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom 14:109
Wang Y, Tang H, DeBarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49
Wang L, Zhao L, Zhang X, Zhang Q, Jia Y, Wang G, Li S, Tian D, Li W, Yang S (2019) Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc Natl Acad Sci 116:18479–18487
Wang Y, Huang L, Li Y, FengZ, MuZ, Wang J, Wu X, Wang B, Lu Z, Li G, Wu X (2022) Checking transformation efficiency for different Lagenaria siceraria genotypes by using seed germination pouches as a growth carrier. Plant Cell Tiss Organ Cult 2022. https://doi.org/10.1007/s11240-022-02345-x
Xu B, Yang Z (2013) PAMLX: a graphical user interface for PAML. Mol Boil Evol 30(12):2723–2724
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci 109:1187–1192
Xue J, Zhao T, Liu Y, Liu Y, Zhang Y, Zhang G, Chen H, Zhou G, Zhang S, Shao Z (2020) Genome-wide analysis of the nucleotide binding site leucine-rich repeat genes of four orchids revealed extremely low numbers of disease resistance genes. Front Genet 10:1286
Yang X, Wang J (2016) Genome-wide analysis of NBS-LRR genes in sorghum genome revealed several events contributing to NBS-LRR gene evolution in grass species. EvolBioinform Online 12:9–21
Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen J, Tian D (2006) Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62:181–193
Yang S, Zhang X, Yue J, Tian D, Chen J (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genom 280(3):187–198
Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q (2015a) Over-expression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol 57:1078–1088
Yang S, Wang L, Huang J, Zhang X, Yuan Y, Chen J, Hurst LD, Tian D (2015b) Parent–progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523:463–467
Yang X, Zhang L, Yang Y, Schmid M, Wang Y (2021) miRNA mediated regulation and interaction between plants and pathogens. Int J Mol Sci 22:2913
Yoshimura S, Yamanouchi U, Katayose Y et al (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci USA 95(4):1663–1668
Yu D, Chen C, Chen Z (2001) Evidence for an important role of wrky DNA binding proteins in the regulation of NPR1 gene expression. The Plant Cell Online 13:1527–1540
Yu J, Tehrim S, Zhang F et al (2014) Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics 15(1):3
Yu X, Feng B, He P, Shan L (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55:109–137
Yu X, Zhong S, Yang H et al (2021) Identification and characterization of NBS resistance genes in Akebia trifoliata. Front Plant Sci 12:758559
Zhang Z, Xie Z, Zou X, Casaretto JA, Ho TD, Shen QJ (2004) A rice wrky gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134:1500–1513
Zhang M, Wu Y, Lee M, Liu Y, Rong Y, Santos TS, Wu C, Xie F, Nelson R, Zhang H (2010) Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Res 38(19):6513–6525
Zhang R, Murat F, Pont C, Langin T, Salse J (2014) Paleo-evolutionary plasticity of plant disease resistance genes. BMC Genomics 15:187
Zhang Y, Xia R, Kuang H, Meyers BC (2016) The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol 33:2692–2705
Zhang Y, Chen M, Sun L, Wang Y, Yin J, Liu J, Sun X, Hang Y (2020) Genome-wide identification and evolutionary analysis of NBS-LRR genes from Dioscorea rotundata. Front Genet 11:484
Zhou T, Wang Y, Chen J, Araki H, Jing Z, Jiang K, Shen JZ, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genom 271(4):402–415
Zhu Q, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I (2013) miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 8:e84390
Funding
This study is financially supported by the Indian Council of Agricultural Research, New Delhi (Grant ID: IXX14668).
Author information
Authors and Affiliations
Contributions
Anuradha Chelliah: project administration, conceptualization, work design, data curation, methodology, visualization, analyzed the data, writing—original draft, review and editing, formatting; Chandrasekar Arumugam: methodology, in silico analysis; Backiyarani Suthanthiram: validation; Thangavelu Raman: validation; Uma Subbaraya: validation.
Corresponding author
Ethics declarations
Ethical approval
No ethical issues involved.
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chelliah, A., Arumugam, C., Suthanthiram, B. et al. Genome-wide identification, characterization, and evolutionary analysis of NBS genes and their association with disease resistance in Musa spp.. Funct Integr Genomics 23, 7 (2023). https://doi.org/10.1007/s10142-022-00925-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-022-00925-w