Abstract
Host genetic factors play an important role in diverse host outcomes after influenza A (H7N9) infection. Studying differential responses of inbred mouse lines with distinct genetic backgrounds to influenza virus infection could substantially increase our understanding of the contributory roles of host genetic factors to disease severity. Here, we utilized an integrated approach of mRNA-seq and miRNA-seq to investigate the transcriptome expression and regulation of host genes in C57BL/6J and DBA/2J mouse strains during influenza virus infection. The differential pathogenicity of influenza virus in C57BL/6J and DBA/2J has been fully demonstrated through immunohistochemical staining, histopathological analyses, and viral replication assessment. A transcriptional molecular signature correlates to differential host response to infection has been uncovered. With the introduction of temporal expression pattern analysis, we demonstrated that host factors responsible for influenza virus replication and host–virus interaction were significantly enriched in genes exhibiting distinct temporal dynamics between different inbred mouse lines. A combination of time-series expression analysis and temporal expression pattern analysis has provided a list of promising candidate genes for future studies. An integrated miRNA regulatory network from both mRNA-seq and miRNA-seq revealed several regulatory modules responsible for regulating host susceptibilities and disease severity. Overall, a comprehensive framework for analyzing host susceptibilities to influenza infection was established by integrating mRNA-seq and miRNA-seq data of inbred mouse lines. This work suggests novel putative molecular targets for therapeutic interventions in seasonal and pandemic influenza.
Similar content being viewed by others
Introduction
Influenza A (H7N9) viruses normally circulate among birds and infect people sporadically. Since the first identification of H7N9 human infection in China in March 2013, a total of 1562 laboratory-confirmed cases have been reported to WHO as of 13 September 2017 (http://www.who.int/csr/don/13-september-2017-ah7n9-china/en/), around 40% of people confirmed with Asian H7N9 virus died. Although the mechanisms by which H7N9 virus infects humans remain unclear (Han et al. 2013; Lam et al. 2013; Xiong et al. 2013; Zhou et al. 2013; Zhu et al. 2013), the idea that host susceptibility plays a significant role is almost certain because only a small fraction of people who are exposed to H7N9 is infected. A list of host genes, such as IFITM3 (Wang et al. 2014), LGALS1 (Chen et al. 2015), and TMPRSS2 (Cheng et al. 2015), has been demonstrated for their roles in regulating host susceptibilities to influenza A (H7N9) virus infection.
For most human diseases, particularly infectious diseases, host genetic susceptibility genes are typically identified using mouse models. Mouse models provide advantages such as availability of large numbers of experimental matings and minimal variation in the environmental influences on disease (Nadeau 2001). Studies on inbred mice have contributed substantially to our understanding of host susceptibility to influenza viruses (Boivin et al. 2012; Boon et al. 2009; Ferris et al. 2013; Nedelko et al. 2012; Toth and Williams 1999). Differential response to influenza A virus infection has been demonstrated in seven inbred mouse strains, among which infected DBA/2J mice exhibited relatively higher susceptibility to influenza infection than infected C57BL/6J mice in terms of higher viral load in the lungs and higher levels of cytokines and chemokines (Srivastava et al. 2009). These findings suggest the major contribution of host genetic background to influenza A virus susceptibility. Another study compared DBA/2J and C57BL/6J mice in terms of their response to H5N1 and identified genetic elements associated with survival (Boon et al. 2009). Both studies indicated that DBA/2J mice exhibit higher susceptibility to influenza infection than C57BL/6J mice. Thus, the comparisons between the two mouse strains can be applied to establish a suitable model system for detecting genes responsible for severe infection outcomes in human. Although several quantitative trait loci (QTL) associated with resistance to H5N1 virus have been identified from genome-wide QTL mapping (Boon et al. 2009; Srivastava et al. 2009), other reports indicated that different QTL regions are associated with response to different influenza virus strains (Bao et al. 2013). Thus, susceptibility to H7N9 might not be determined by these previously identified genes that affect the infectivity of other strains. The identification of H7N9 susceptibility genes is an urgent and challenging task.
During acute influenza A virus infection, highly dynamic and inter-related responses are triggered in the host, resulting in clearance of pathogens and establishment of long-lasting immunity. Several transcriptome studies suggested that host responses to influenza infection could be studied comprehensively by measuring changes in gene expression levels after infection (Alberts et al. 2011; Pommerenke et al. 2012). On the other hand, recent research has indicated that virus infection can induce the expression of cellular microRNA (miRNA) species that modulate the stability of their target host mRNAs and hence their protein products.
In this study, we used next-generation sequencing (NGS) technology to characterize mRNA and miRNA levels in C57BL/6J and DBA/2J mice before and after virus infection to elucidate the molecular mechanisms of host susceptibility. We found that influenza A (H7N9) virus exhibits great differential pathogenicity between C57BL/6J and DBA/2J mice. The global gene expression profiles of these two mouse strains were distinct. Through the combination of time-series expression level analysis and temporal expression pattern analysis (i.e., DTW-MIC (Riccadonna et al. 2016)), we identified a list of promising candidate genes and pathways that may play pro-/anti-influenza roles after infection. A synergistic miRNA regulatory network responsible for host susceptibility was constructed using Mirsynergy (Li et al. 2014) by integrating all candidate genes identified, the genomic signatures embedded in mRNA-seq and miRNA-seq data, and the functional knowledge of protein–protein interactions and miRNA–target prediction. These results can help us understand more about the complex mechanisms underlying the susceptibility and resistance of genetically diverse hosts to influenza infection.
Results
Differential pathogenicity of H7N9 in C57BL/6J and DBA/2J
We detected a higher amount of H7N9 antigens in infected DBA/2J mice than that in infected C57BL/6J mice under the same infection conditions (Fig. 1a). The results are highly consistent with the higher amount of viral RNAs detected in infected DBA/2J mice than that in infected C57BL/6J mice (Additional File 1, Supplementary Fig. S1). Histopathologically, we detected significant peribronchiolar interstitial infiltration, bronchiole epithelial cell necrosis, and necrotic cell debris within the alveolar lumens of the H7N9-inoculated DBA/2J lungs (Fig. 1b). Comparatively, mild peribronchiolar interstitial infiltration was observed in H7N9-inoculated DBA/2J lungs. Similar results were obtained from RNAseq analysis of 10 interferon-stimulated genes (including CXCL10, IFIT1, IFIT3, IFNA4, IFNB1, IL1B, ISG15, MX1, RSAD2, and USP18) (Chen et al. 2016). All of these interferon-stimulated genes were upregulated after H7N9 infection, and their differential expression levels in DBA/2J were higher than that in C57BL/6J after infection for 3 days (Additional File 1, Supplementary Fig. S2). Our results demonstrated that DBA/2J was more susceptible to influenza A (H7N9) virus infection than C57BL/6J, consistent with the findings of previous studies (Boon et al. 2009; Srivastava et al. 2009).
C57BL/6J and DBA/2J show distinct phenotypes after influenza A (H7N9) virus infection. a Immunohistochemical staining of H7N9 antigens in virus-inoculated DBA/2J (D2) and C57BL/6J (B6) lungs. Representative images of immunohistochemically stained influenza nucleoprotein (NP) in formalin fixed mouse lung tissue infected with H7N9 at day 3 post-infection (right panels). Viral NP protein was labeled brown by 3,39-diaminobenzidine and then countered with hematoxylin. Uninfected mouse lung as negative control (left panels). Images were taken at magnification of × 200. b Histopathological changes in the D2 and B6 lung tissues infected with H7N9. Representative histological images of hematoxylin and eosin (H&E)-stained lung tissue sections of normal mouse lung (left panels) and infected mouse lung (right panels) at day 3 post infection. Images were taken at magnification of × 100
Genomic signatures embedded in RNAseq data reveal distinct host response to influenza infection and virus temporal dynamics
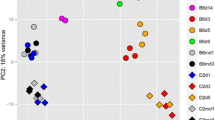
Differential host responses to influenza infection between susceptible and resistant hosts exhibit a distinct temporal molecular signature. To pick up this genomic signature in an unbiased manner, we analyzed the global expression profile by using an unsupervised method called non-negative matrix factorization (NMF) (Dhillon and Sra 2005; Tandon and Sra 2010); this method requires no prior knowledge of phenotype information, such as sample or time label and disease outcome. The entire gene expression profile was clustered into a predefined number (i.e., 4) of groups. To promote robust cluster assignments, we obtained a consensus clustering assignment after aggregating the results of 200 different runs into a single clustering result. Figure 2a shows that all samples were grouped into three main clusters, except C57-D3-3 (which refers to C57BL/6J mice on day 3 after infection, biological replicate no. 3) that was grouped as a single cluster. After introducing sample information about strains and conditions (infected or uninfected) into the consensus matrix, we found that all C57BL/6J samples (i.e., resistant mice), except C57-D3-3, were grouped together. Uninfected DBA/2J mice and infected DBA/2J mice on day 1 (i.e., susceptible mice at early-stage of infection) were grouped into another cluster. Infected DBA/2J mice on days 2 and 3 (i.e., susceptible mice at the late stage of infection) were grouped as the third cluster. From the distribution of the amount of viral RNAs in each cluster (Fig. 2b), we found that cluster 3 possessed higher load of virus than clusters 1 and 2. This finding demonstrated the strength of NMF for ab initio discovery of genomic signature responsible for host response to influenza infection. Principle component analysis (PCA) of the normalized host gene counts for all samples also confirmed the separate groupings of mice at early and late stage of infection (Fig. 2c). The diverse host response to influenza infection was mainly represented by the first principle component (i.e., PC1), which explained 42% of the expression variation. PC2 revealed distinct expression profiles of C57BL/6J and DBA/2J due to their different genetic backgrounds and explained around 17% of the total expression variation.
A genomic signature shows distinct host response to infection. a Consensus matrix (average connectivity matrix) of samples across 200 runs. The consensus score assigned for a given pair of samples was calculated as the percentage of assignments that these two samples were grouped together among all clustering runs, and was represented as heating colors from cold blue (score = 0) to hot read (score = 1). Sample labels for infected mice were stated as in “strain-day time-biological replicate” format, i.e. C57-D1-1 represents an infected C57BL/6J mouse (biological replicate 1) at day 1. For mock-treated mice, “C” character was added before the biological replicate number, i.e., C57-D1-C1 represents a mock-treated (control) C57BL/6J mouse (biological replicate 1) at day 1. Different conditions (infected and mock-treated) and strains (C57BL/6J and DBA/2J) were represented by different groups of colors as shown in the legend. b The distribution of virus load in three main clusters. c PCA of normalized host gene counts of all samples. Replicates for a given day which were represented by the same color were grouped together well. Different strains were also stratified well by the second principle component
In addition to the genomic signatures of host genes, RNAseq data also allowed us to analyze the expression profile of eight viral segments (i.e., M2, NA, HA, NP, NEP, PB2, PB1, PA). After infection, the overall expression levels of influenza transcripts were higher in DBA/2J mice than those in C57BL/6J mice from day 1 to day 3. The global expression levels of all influenza segments were correlated well (Pearson correlation = 0.48, p value = 0.005) with the amount of viral RNAs (Fig. 3).
Dynamics of influenza gene expression level correlated with the amount of viral RNAs in C57BL/6J and DBA/2J. a Expression levels of influenza segments changed in infected mice compared with mock-treated mice. The expression levels of influenza segments changed over time were calculated as the mean expression value of (log2RPKM + 1) in infected mice at the same day, relative to that in respective mock-treated mice (mock day 1 for day 1 and day 2 infected mice, mock day 3 for day 3 infected mice). Lines represent expression levels from lungs of C57BL/6J (B6d1, B6d2, B6d3) and DBA/2J at day 1, 2, and 3 (D2d1, D2d2, D2d3). b The amount of viral RNAs extracted from the lungs of mock-treated mice and infected mice at day 1, 2, and 3
Host factors differentially expressed between strains either in terms of overall expression level or temporal expression pattern
To fully use the time point data available, we used mock-treated mice on days 1 and 3 as infected mice on day 0 (before infection) and carried out time-series analysis of differentially expressed genes (DEGs) between strains over time by logistic regression test in DESeq2 (Love et al. 2014). A total of 224 genes were significantly differentially expressed between C57BL/6J and DBA/2J mice over time and defined as strain-specific DEGs (Additional File 2, Supplementary Table S1). The transcriptome profiles of these DEGs showed distinct expression patterns between susceptible and resistant mice (Additional File 1, Supplementary Fig. S3 A), and around 26% of total expression variation of these genes can be explained by genetic background (Additional File 1, Supplementary Fig. S3 B).
Likelihood ratio tests can help us identify genes exhibiting differential expression levels between strains over time. However, a group of genes may have insufficient time to show obvious differential expression levels within a short period of time but exhibit distinct temporal expression patterns in different hosts. This type of genes should also play a role in host susceptibility to influenza infection. To measure the similarity of a gene between C57BL/6J and DBA/2J in terms of temporal expression pattern, we employed a similarity function called dynamic time warping maximal information coefficient (DTW-MIC) (Riccadonna et al. 2016); this function combined a similarity measure taking care of non-linear interactions (MIC) and a measure identifying time shifts (DTW) (Riccadonna et al. 2016). Around 18.14 and 39.75% of all genes with meaningful DTW-MIC scores (21,554 genes; Supplementary Table S2, S3) were enriched in DTW-MIC similarity ranges of 0.2–0.25 and 0.7–0.75, respectively (Additional File 1, Supplementary Fig. S4 A). We then carried out permutations by randomly swapping the phenotype labels of C57BL/6J and DBA/2J 10,000 times. In each permutation, we calculated the permutated DTW-MIC scores for all genes. Genes with DTW-MIC scores lower than 0.25 achieved FDR less than 0.05 (the real DTW-MIC score was lower than 95% of all corresponding permutated scores; Additional File 1, Supplementary Table S3), leaving a set of (3910) genes with significantly low expression similarities between C57BL/6J and DBA/2J mice, termed as sig.low.simi genes for brevity. The expression variation across samples from this set of genes was mainly attributed to host genetic background, which explained around 46% of the total expression variation (Additional File 1, Supplementary Fig. S4 B). By contrast, genes with DTW-MIC scores higher than 0.25 mainly explained the host response to influenza infection, and this characteristic was the most obvious among genes with DTW-MIC scores between 0.7 and 0.75, where the PCA showed the most separated groupings of samples per host response to influenza infection.
We then mapped the two list of candidate genes, namely, strain-specific DEGs and sig.low.simi genes, responsible for host susceptibility to a list of genes that were reported by at least two RNAi screens for their roles in influenza virus replication (Chou et al. 2015) (Additional File 2, Supplementary Table S4) and a list of host proteins that were reported as interacting with influenza virus (Shapira et al. 2009) (Additional File 2, Supplementary Table S5). No strain-specific DEGs were reported as host factors responsible for viral replication and/or host-virus interaction, whereas mouse transcripts homologous to those host factors were significantly enriched in genes with significantly low temporal similarities between strains (Additional File 2, Supplementary Table S6). Specifically, 33 out of 3910 sig.low.simi genes (hypergeometric distribution p value = 0.031) were mapped to host factors associated with influenza virus replication, and 22 out of 3910 genes (hypergeometric distribution p value = 0.022) were reported to interact with the influenza virus. This result demonstrated that a significant amount of host factors responsible for viral replication and/or host-virus interaction show distinct temporal dynamics in different hosts instead of differential expression levels.
A total of 82 genes were identified as promising candidates for future studies through differential expression level analysis and temporal expression pattern analysis (Additional File 2, Supplementary Table S7). Among them, A2M was reported as a natural inhibitor against influenza A virus of swine origin (Chen et al. 2010; Ryan-Poirier and Kawaoka 1993). Gpx2 is an important member of glutathione peroxidases subfamily of selenium; adequate selenium intake plays a vital role in influenza A (H3N2) virus clearance and recovery (Beck et al. 2001), and supplementation of this element protects against influenza A (H1N1) infection (Yu et al. 2011). In line with this, Gpx2 was significantly downregulated in DBA/2J after infection, and the level of Gpx2 in C57BL/6J exhibited almost no change (Additional File 2, Supplementary Table S7), suggesting that the expression level of Gpx2 in host cells may also play an important role in anti-influenza severity when infected by influenza A (H7N9) virus. Pla2g16 was exclusively upregulated in DBA/2J mice and may play a pro-H7N9 role in facilitating viral genome translocation and preventing viral clearance (Staring et al. 2017). Functionally, the overlapped candidate genes from time-series expression level analysis and temporal expression pattern analysis were mainly enriched in immune system and apoptotic process (Additional File 2, Supplementary Table S8), indicating that these two processes may respond distinctly not only in terms of differential expression levels but also in terms of diverse dynamics in different hosts after being infected by influenza virus.
Hosts with distinct genetic background tend to participate in different biological processes after influenza infection
We hypothesized that genes that show distinct temporal dynamics between susceptible and resistant strains and also differentially express between infected and uninfected mice in either one of the two strains are more likely responsible for host susceptibility to influenza than those which did not differentially express in either of the two strains. Thus, we divided all sig.low.simi genes into genes that were significantly differentially expressed between infected and uninfected mice over time in both C57BL/6J and DBA/2J strains (cluster 1), exclusively in C57BL/6J strain (cluster 2), exclusively in DBA/2J strain (cluster 3), and in neither of these two strains (cluster 4; Additional File 2, Supplementary Table S10). Pathway enrichment analysis showed that cluster 1 genes were significantly enriched in negative regulation of cardiac muscle cell proliferation, whereas genes from cluster 2 and 3 show distinct pathway enrichments between each other, except for a common link to the modulation of host immune and inflammatory response (Table 1). Similar observations were also obtained from the analysis of protein–protein interaction networks in all genes from clusters 1, 2, and 3, where genes from clusters 2 and 3 tended to aggregate within clusters and formed several cluster-specific functional modules, such as oxidative stress, ribosome, and cell cycle (Fig 4). There are two possible explanations for this observation: first, besides immune-related pathways, the influenza virus may trigger different biological processes in hosts with different genetic background; and second, different hosts may have their own tendencies to participate in one or several mechanisms in response to influenza infection, which finally led to their resistance or susceptibility after being infected by influenza.
Interaction network of genes with significantly low temporal similarities between C57BL/6J and DBA/2J and differentially expressed in at least one strain. Genes from different clusters were represented by different colors as shown in the legend. Aggregated modules of genes were grouped by dashed circles in different colors. Representative pathways enriched by a certain module of genes were shown at the top of each corresponding enlarged drawings. More details about member genes and pathway enrichment of each module can be found in S11 Table
Cell cycle and ribosome in pro-/anti-influenza infection
Cell cycle and ribosome were the two pathways specifically enriched in and aggregated by genes that were exclusively differentially expressed in C57BL/6J (cluster 2) and DBA/2J (cluster 3), respectively. Influenza A virus in infected cells induces a pro-influenza environment by arresting a G0/G1-phase cell cycle(Fan et al. 2017; He et al. 2010; Jiang et al. 2013) to promote viral protein accumulation and virus production (He et al. 2010) or inhibit early cell death of infected cells, allowing the cells to evade immune defenses (Bagga and Bouchard 2014). Delayed mitotic exit has also been implicated in the pathogenesis of viral infection (Heilman et al. 2005). The upregulation of some cell cycle molecules opposing the completion of mitosis was linked to disease severity associated with influenza infection (Parnell et al. 2011). By contrast, the dysregulation of the anaphase promoting complex (APC), a major regulatory complex which coordinates cell cycle progression and mitotic exit(Castro et al. 2005), was found in subjects with the most severe infection (Parnell et al. 2011). Knockdown of cyclin D3, a key regulator of cell cycle in the early G1 phase, led to cell cycle G0/G1 phase arrest and significantly enhanced influenza virus replication (Fan et al. 2017). In summary, host response to influenza infection can be characterized by opposing changes in cell cycle activity (e.g., G0/G1 phase arrest and/or delayed mitotic exit). In line with this, genes that promote cell cycle progression and exit from mitosis were exclusively significantly upregulated in resistant C57BL/6J mice but not in susceptible DBA/2J mice; these genes include cell cycle regulator (CCNA2), a trigger of mitotic entry (CDCA3), a component of the mitotic checkpoint (KNTC1), and the mitotic-specific ubiquitin-conjugating enzyme (UBE2C) (Additional File 1, Supplementary Fig. S5; Additional File 3), indicating their potential anti-influenza role in protecting hosts from highly lethal infection.
Viruses recruit host ribosomes to translate viral mRNAs, which not only ensures that viral proteins are produced but also stifles innate host defenses to inhibit the capacity of infected cells for protein synthesis (Walsh and Mohr 2011). In influenza-infected cells, some mRNAs for ribosomal proteins, such as S6 (Dufner and Thomas 1999) and RRS1 (Tsuno et al. 2000), were found to be highly upregulated, despite an overall inhibition of the synthesis of infected cellular proteins (Geiss et al. 2001). Not all genes involved in protein synthesis were upregulated, indicating that the influenza virus may recruit specific components for viral mRNA translation (Geiss et al. 2001). Since host ribosome regulates viral protein synthesis, a worth trying in discovering new potential therapeutic target will be focusing on the protein components on the surface of the ribosome. The blockade of RPL40 disables vesicular stomatitis virus cap-dependent translation while leaving normal cells largely unaffected, demonstrating that besides catalyzing the synthesis of cellular proteins, ribosomes may act as a translational regulator (Lee et al. 2013). Although no direct links between ribosomal proteins and influenza A virus translation have yet been established, significant perturbations in host proteome were found very early in influenza A (H5N1) virus-infected human macrophages; the most pronounced effect was observed on the translational machinery (Cheung et al. 2012). Several member proteins of the small 40S subunit, including RPS5, RPS18, and RPS25, which were reported to be significantly upregulated post-infection in H5N1 macrophages (Cheung et al. 2012), exhibited higher significant differential expression levels between infected and uninfected mice in DBA/2J strain than that in C57BL/6J strain (Additional File 1, Supplementary Fig. S6 A; Additional File 3), indicating their potential pro-influenza role in influenza virus replication. In addition, three other genes encoding ribosomal proteins, including RPS14 (Hao et al. 2008; Karlas et al. 2010), RPL15 (Hao et al. 2008), and RPL13A (Brass et al. 2009; Shapira et al. 2009), which were reported to interact with viral ribonucleoproteins and are involved in influenza virus life cycle, showed much more significant upregulation in DBA/2J than in C57BL/6J (Additional File 1, Supplementary Fig. S6 B, C, D; Additional File 3). These results provide new insights into the role of host ribosome in the pathogenicity of influenza.
Integrated miRNA regulatory network responsible for host susceptibilities
In addition to RNA-seq data, we also extracted miRNAs from the lungs of the same series of mice and performed miRNA-seq. A miRNA regulatory network responsible for host susceptibilities was constructed by integrated analysis of RNA-seq data and miRNA-seq data through Mirsynergy(Li et al. 2014)—a deterministic overlapping clustering algorithm that expands miRNA-mRNA module by greedily including mRNAs into the module to maximize the synergy score, which is defined as a function of the miRNA-mRNA interaction weight (MMIW) matrix and the gene–gene interaction weight (GGIW) matrix. All strain-specific DEGs over time and genes with significantly low similarities between strains were collected as input genes. miRNAs which were predicated as the regulators (context score <− 0.2) of these candidate genes by TargetScan v7.1(Agarwal et al. 2015) were also included. The MMIW matrix was determined by expression-based correlations between miRNAs and corresponding mRNAs using L1-norm regularized linear regression model (i.e., LASSO). The binary interaction weight matrix between mRNAs (i.e., GGIW matrix) was determined by STRING v10 (Szklarczyk et al. 2015) association scores higher than 400 (medium confidence). A total of 28 regulatory modules (Additional File 4) were constructed. For simplification, we combined modules with over half of members overlapping together as a group, leaving behind 13 groups (Fig. 5; Additional File 4).
Synergistic miRNA regulatory network responsible for host susceptibilities to influenza infection. Genes were represented by circles while miRNAs were shown as diamonds. The interactions between miRNAs and genes were highlighted by blue, and the corresponding target genes were circled by blue. Gene and/or miRNAs from different groups were covered by solid circles in different colors as shown in the legend. Most significant and representative pathways enriched by each group of genes were listed beside the pathway-involved genes
Functional enrichment analysis of these groups indicated the roles of several pathways in regulating host susceptibilities to influenza infection, including MAPK signaling pathway (Gaur et al. 2011), PI3K/Akt pathway (Ehrhardt et al. 2007; Zhao et al. 2014), endocytosis (Fujioka et al. 2011; Lakadamyali et al. 2004; Sieczkarski and Whittaker 2003), ribosome (Cheung et al. 2012; Lee et al. 2013; Walsh and Mohr 2011), and calcium signaling pathway (Fujioka et al. 2013). The MAP kinase (MAPK) Rac1/p38 plays a key role in regulating inflammatory responses (Mainiero et al. 2000). Inhibition of Rac1 by NSC23766 leads to impaired replication of influenza virus (Dierkes et al. 2014). Three miRNAs, including miR-302c, miR-200, and miR-320, were predicted to target this gene, and how Rac1 is regulated in different hosts is worth investigating. Bcl2, which is the direct neighbor of Rac1, is an apoptosis regulator, is involved in PI3K/Akt signaling pathway, and exhibits an anti-influenza role in both reduced virus-induced apoptosis [66, 67] and viral replication [67, 68]. Intriguingly, both Rac1 and Bcl2 were neither differentially expressed between strains nor differentially expressed between infected and uninfected mice in either of the two strains but showed distinct expression patterns between strains (Additional File 2, Supplementary Table S9).
The integrated regulatory network also revealed several important miRNAs contributing to anti-influenza virus. MiR-491 inhibits H1N1 replication and was predicted to complement the 3’-UTR of Tns4 mRNA, which is involved in apoptosis. MiR-34a inhibits influenza virus-induced apoptosis (Fan and Wang 2016). The predicted target of miR-34a, that is, Chmp7, functions in endocytosis.
Discussion
Combination of expression level analysis and temporal expression pattern analysis
As a classical gene expression analysis strategy, the likelihood ratio test can reveal the overall contrast of a gene between two conditions in terms of differential expression level. However, a set of disease genes may have insufficient time to show distinct expression level difference between conditions within the period of study. To reduce false negatives from pure differential expression level analysis, we introduced a temporal expression pattern analysis to help us capture genes with distinct temporal dynamics between conditions within the observation period despite the low differential expression level. In the present study, among the genes which show no obvious expression level differences between C57BL/6J and DBA/2J but exhibit distinct expression pattern between these two strains (accounting for over 96% of total sig.low.simi genes; Additional File 2, Supplementary Table S9), Rac1(Dierkes et al. 2014), and Bcl2 (Nencioni et al. 2009; Nencioni et al. 2003; Olsen et al. 1996) have been demonstrated for their pro-/anti-influenza role as mentioned above. Cd1d−/− mice was shown to have higher IAV and reduced PR8-specific CD8+ T lymphocytes and antibodies compared with PR8-infected wide type mice (De Santo et al. 2008), suggesting the anti-influenza role of Cd1d. Furthermore, with the introduction of time course expression pattern analysis, we have noticed the roles of a list of novel pathways, such as cell cycle and ribosome, in host susceptibilities to influenza infection.
Genes differentially expressed between C57BL/6J and DBA/2J mice over time do not necessarily show distinct temporal dynamics between these two strains at the same time (Additional File 1, Supplementary Fig. S7). Over 57% (129 out of 224) of all strain-specific DEGs had DTW-MIC similarity scores higher than 0.7 (Additional File 1, Supplementary Fig. S7 A). The expression variation across samples based on these genes were mostly from host response to influenza infection (Additional File 1, Supplementary Fig. S7 B and C, right inset), indicating that a high false positive rate may exist in differential expression analysis.
Although a systematic evaluation of the sensitivity and specificity of differential expression level analysis and temporal expression pattern analysis is yet to be performed, the introduction of DTW-MIC similarity function in RNA-seq expression data analysis has provided us a new direction in measuring the difference of a gene between two conditions. Moreover, a combination of candidate genes from both expression level analysis and expression pattern analysis can provide a more comprehensive understanding of host factors responsible for different outcomes after influenza infection.
Integration of multiple layers of information
The major part (over 87%; Additional File 2, Supplementary Table S10) of the sig.low.simi genes were from genes differentially expressed between infected and uninfected mice in either of the two strains (cluster 4). With the advancement of a miRNA regulatory network and literature support, we have uncovered several promising candidates responsible for host susceptibilities, including Rac1 (Dierkes et al. 2014) and Bcl2 (Nencioni et al. 2009; Nencioni et al. 2003; Olsen et al. 1996), which have been reported for their anti-/pro-influenza roles in previous studies. However, a great number of sig.low.simi genes, especially those that were neither differentially expressed between infected and uninfected, nor correlated with virus load (which account for around 61.5% of total sig.low.simi genes), have no further information to explain their roles in influenza pathogenesis in our current data. Integrating additional layers of information beyond transcriptome expression and miRNA regulation, such as genotype information, epigenome, copy number variations, and so on, into a composite analysis framework may further expand our ability to pinpoint the host factors responsible for different outcomes after influenza infection.
Conclusions
Immunohistochemical and histopathological analyses indicated that the two mouse lines (i.e., C57BL/6J and DBA/2J) show distinct susceptibilities upon being infected with influenza virus. The genomic signatures in RNA-seq data of these two mouse lines are so strong that we can group all biological samples according to their response to influenza infection through a clinically uninformed factor analysis method (i.e., NMF). By introducing temporal sequence similarity measurement (i.e., DTW-MIC), we identified a list of genes which show distinct temporal expression patterns between C57BL/6J and DBA/2J, called sig.low.simi genes. Intriguingly, host factors which have been suggested for their roles in influenza virus replication and host-virus interactions were significantly enriched in the sig.low.simi genes. A combination of both traditional time-series gene expression level analysis and temporal expression pattern analysis revealed that hosts with different genetic background may behave distinctly in immune systems and apoptotic process when infected by influenza infection not only in terms of overall expression level but also in terms of temporal dynamics. In addition, we suggest that susceptible and resistant hosts may have their own preference in triggering internal biological processes in response to influenza infection. Several pathways, such as cell cycle and ribosome, which are yet to be well-characterized for their roles in influenza pathogenesis, may play a much important role in pro-/anti-influenza infection than we have ever expected. Furthermore, a miRNA regulatory network formed by integrating both RNA-seq data and miRNA-seq data of the two mouse lines has revealed several regulatory modules (e.g., MAPK signaling, ubiquitin, endocytosis, ribosome, and calcium signaling), which were responsible for host susceptibility, and a list of miRNAs (e.g., miR-491 and miR-34a), which have been reported for their roles in anti-/pro-influenza virus. The predicted target genes of these miRNAs in the network add important knowledge to miRNA-gene regulation in directing diverse host outcomes after influenza infection. Taken together, the results provided here will advance our understanding of the mechanisms underlying host susceptibilities or survival during influenza A (H7N9) infection.
Methods
Inbred mouse strains
C57BL/6J mice were obtained from the Animal Unit of the University of Hong Kong. DBA/2J mice and BXD recombinant inbred mice derived from crosses between C57BL/6J and DBA/2J were imported from the Jackson Laboratory.
Infection of mice with H7N9 virus
Mice were infected with human H7N9 strain isolated from Shenzhen/HK in 2014 and kept by Department of Microbiology, HKU and State Key Laboratory for Emerging Infectious Disease. Infection was done in P3 lab in the Department of Microbiology, HKU. Experimental procedures have been described (Xiong et al. 2013). In brief, mice were anesthetized and virus was administered intra-nasally. Afterward, we harvested the lungs from infected mice on days 1, 2, and 3 and collected the lungs from mock-treated mice on days 1 and 3. For each strain at days 1 and 3, four infected mice and four mock-treated mice were used for biological replication. We only harvested the lungs of four infected mice on day 2, assuming that mock-treated mice should have very little variation in terms of gene expression from day 1 to day 2. Harvested lungs were stored at 80 °C. We pooled lung RNAs or microRNAs from the same mouse at each time point. Pooled RNAs or microRNAs were sequenced in duplicate. The lethal dose of H7N9 in C57BL/6J and DBA/2J mice was determined as previously described (Boon et al. 2009).
Immunohistochemical staining, histopathological analyses, and viral replication assessment of infected mouse lung tissues
Infected mouse lungs were fixed in 10% paraformaldehyde and paraffinized. After mounting on slides, the tissues were de-paraffinized, rehydrated, and treated with Antigen Unmasking Solution according the manufacturer’s instructions (Vector Laboratories Inc. Burlingame, CA, USA) to unmask the antigens. The influenza virus nucleoprotein (NP) was then stained with mouse anti-influenza NP-antibody (HB65, ATCC) at 4 °C for overnight after blocking with 3% bovine serum albumin as previously described (Yeung et al. 2016). After washing, biotin-conjugated goat anti-mouse IgG (Calbiochem, Darmstadt, Germany) was added to the tissue sections at room temperature for 30 min. Streptavidin/peroxidase complex reagent (Vector Laboratories, Burlingame, CA) was then added and incubated at room temperature for 30 min after washing. Color development was allowed after the addition of substrates 3,39-diaminobenzidine (DAB, Vector Laboratories, Burlingame, CA, USA). Digital images were acquired using NIKON Eclipse Ni-U with SPOT RT3 camera. Histopathological changes of the infected mouse lung tissues were examined by staining with hematoxylin and eosin after deparaffinization as described above. Viral replication and virus shedding were determined by quantitative RT-PCR, plaque formation, and influenza hemagglutination inhibition assay.
Gene expression analysis of H7N9-infected mice by NGS (RNA-Seq)
All procedures were performed following the manufacturer’s instructions. In brief, total RNA was extracted by Rneasy (Qiagen). RNA quality was checked using the Bioanalyzer 2100 (Palo Alto, CA). cDNA was synthesized using the Superscript system (InVitrogen) and the Enzo BioArray High Yield RNA transcript Labeling kit (Enzo Biochem). Purified cRNA was sequenced in an Illumina HiSeq2000 Analyzer at the Centre for Genomic Sciences of the University of Hong Kong.
miRNA analysis of H7N9-infected mice by NGS (miRNA-Seq)
RNA samples were prepared using the DGE-Small RNA Sample Prep Kit (Illumina, San Diego, CA). Small RNAs of 18–30 nt in length were prepared from total RNA by gel fractionation and then ligation to linkers. cDNA libraries were generated by RT-PCR and then amplified with 15 PCR cycles to produce sequencing libraries. Purified cDNAs were quantified using the QuantiT PicoGreen dsDNA Kit (Invitrogen, Carlsbad, CA) and diluted to 10 nM for sequencing on an Illumina HiSeq2000 Analyzer at the Centre for Genomic Sciences of the University of Hong Kong.
Statistical analysis
NMF method was used to detect genomic signature without the supervision of prior class information (Fig. 2a). Similar to other unsupervised factorization methods, NMF finds a decomposition of the large input matrix V into two matrices: one of metagenes matrix (W) and the other of metagene expression profiles (H). Specifically, let V be a n × p non-negative matrix, (i.e., with Vij ≥ 0, denoted V ≥ 0), and r > 0 an integer. NMF consists in finding an approximation: X ≈ WH, where W, H are n × r and r × p (r < < min (n, p)) non-negative matrices, respectively. Intuitively, an NMF-discovered metagene can be viewed as a gene expression profile, whose amplitudes represent the relative contribution of each gene to that metagene, and the metagene loadings are the proportions of these metagenes that are present in the whole gene expression profile. Such positive constraints used in this method are natural for RNAseq data because the RNAseq read count data is always non-negative. In this application, NMF was run on the whole gene expression profile in terms of normalized gene read count, and extracted a total of four major NMF metagenes. Multiple runs were applied to achieve stability because the seeding method for NMF initialization is stochastic. The final sample assignment was obtained from the consensus matrix (i.e., average connectivity matrix across the runs) after 200 runs.
Raw read counts from mRNA-seq and miRNA-seq data were taken as inputs for DESeq2(Love et al. 2014). The DESeq2 function “rlog Transformation” was used to normalize and log transform raw read counts. The normalized expression counts were then used for further analysis without applying any additional pre-processing filtering. Heatmaps of sample-to-sample Euclidean distance and principle component analysis of normalized gene counts were constructed using DESeq2. For each mouse strain, we carried out time-series analysis for genes differentially expressed between infected and uninfected mice over time by logistic regression test in DESeq2, with mock-treated mice at day 1 used as control data for the infected mice at day 2. Genes differentially expressed between strains over time were detected by logistic regression in DESeq2. Note that we grouped mock-treated mice at day 1 and day 3 as infected mice at day 0 (before infection) to increase the time points. In both of these time-series expression level analyses, we defined DEGs as genes with adjusted p value < 0.05 (FDR of 5%) and exhibiting at least twofold difference in expression levels (|log2-fold change| > 1). All mRNA-seq sequence reads were also aligned to eight influenza gene segments. The sum of normalized log2-tranformed RPKM (log2RPKM + 1) values of all influenza segments was used as influenza gene expression values and correlated with influenza virus loads (TCID50) by Pearson correlation test.
We used DTW-MIC method (Riccadonna et al. 2016) for temporal expression pattern analysis. For a given gene, the expression value expressed in each strain (C57BL/6J or DBA/2J) at a given day (day 0, 1, 2 or 3) was calculated as the median expression value of the biological replicates from the same strain at the same day. DTW-MIC method was used to measure the similarity of the expression patterns between two strains for each gene. Note that genes with read count covered on none of the samples of one strain or covered on too few samples may lead to no meaningful DTW-MIC scores; thus, these genes were excluded for further analysis. Permutations were carried out by randomly swapping strain labels of C57BL/6J and DBA/2J for both infected and mock-treated mice for 10,000 times. Each gene with available DTW-MIC similarity score was assigned a permutation p value by calculating the proportions of DTW-MIC scores under permutations lower than real DTW-MIC score. Genes with significantly low (DTW-MIC score < 0.25, permutation p value < 0.05) similarities between strains were collected and stated as sig.low.simi genes. Among those sig.low.simi genes, genes which were also differentially expressed between infected and uninfected mice over time in at least one strain were grouped into three clusters according to in which strain(s) the gene was differentially expressed after infection. All gene members of these three clusters were collected for functional enrichment analysis and interaction network analysis. Specifically, the interaction network of these cluster genes was constructed by using the interaction scores from STRING v10, whereby only interaction scores higher than medium confidence (score > 400) were included. MCL clustering embed in the AutoAnnotate application of Cytoscape (Shannon et al. 2003) was used to aggregate the interaction network, with edge weighted by the interaction scores between nodes. Aggregated modules with nodes more than 10 were selected for functional enrichment analysis.
All sig.low.simi genes and DEGs between strains over time were collected as candidate genes responsible for host susceptibilities to influenza infection. An integrated miRNA regulatory network was constructed by using all candidate genes and their corresponding regulatory miRNAs by Mirsynergy (Li et al. 2014). The miRNA-mRNA target prediction was carried out by TargetScan v7.1 (Agarwal et al. 2015). The interaction matrix between miRNA and mRNA (MMIW) was constructed according to the expression level-based correlations between miRNAs and their corresponding target genes. L1-norm regularized linear regression model (i.e., LASSO) was used to calculate the correlations. The binary interaction matrix between genes (GGIW) was determined by the interaction scores with at least medium confidence in STRING v10. Synergistic modules produced by Mirsynergy were grouped together if over half of their members (either miRNA or gene) were overlapped. KEGG pathway enrichment analysis of each group of genes was carried out by Enrichr (Chen et al. 2013; Kuleshov et al. 2016).
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NMF :
-
non-negative matrix factorization
- PCA :
-
principle component analysis
- DEGs :
-
differentially expressed genes
- DTW-MIC :
-
dynamic time warping maximal information coefficient
- MMIW :
-
miRNA-mRNA interaction weight
- GGIW matrix :
-
gene–gene interaction weight
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:4. https://doi.org/10.7554/eLife.05005
Alberts R, Lu L, Williams RW, Schughart K (2011) Genome-wide analysis of the mouse lung transcriptome reveals novel molecular gene interaction networks and cell-specific expression signatures. Respir Res 12:61. https://doi.org/10.1186/1465-9921-12-61
Bagga S, Bouchard MJ (2014) Cell cycle regulation during viral infection Methods. Mol Biol 1170:165–227. https://doi.org/10.1007/978-1-4939-0888-2_10
Bao S, Zhou X, Zhang L, Zhou J, To KKW, Wang B, Wang L, Zhang X, Song YQ (2013) Prioritizing genes responsible for host resistance to influenza using network approaches. BMC Genomics 14:816. https://doi.org/10.1186/1471-2164-14-816
Beck MA et al (2001) Selenium deficiency increases the pathology of an influenza virus infection. FASEB J 15:1481–1483
Boivin GA, Pothlichet J, Skamene E, Brown EG, Loredo-Osti JC, Sladek R, Vidal SM (2012) Mapping of clinical and expression quantitative trait loci in a sex-dependent effect of host susceptibility to mouse-adapted influenza H3N2/HK/1/68. J Immunol 188:3949–3960. https://doi.org/10.4049/jimmunol.1103320
Boon AC et al (2009) Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J Virol 83:10417–10426. https://doi.org/10.1128/JVI.00514-09
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile Virus, and Dengue Virus. Cell 139:1243–1254. https://doi.org/10.1016/j.cell.2009.12.017
Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T (2005) The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 24:314–325. https://doi.org/10.1038/sj.onc.1207973
Chen CH et al (2010) The essentiality of alpha-2-macroglobulin in human salivary innate immunity against new H1N1 swine origin influenza A virus. Proteomics 10:2396–2401. https://doi.org/10.1002/pmic.200900775
Chen EY et al (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. https://doi.org/10.1186/1471-2105-14-128
Chen M et al (2016) TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell 64:105–119. https://doi.org/10.1016/j.molcel.2016.08.025
Chen Y et al (2015) Functional variants regulating LGALS1 (galectin 1) expression affect human susceptibility to influenza A(H7N9). Sci Rep 5:8517. https://doi.org/10.1038/srep08517
Cheng ZS et al (2015) Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J Infect Dis 212:1214–1221. https://doi.org/10.1093/infdis/jiv246
Cheung CY et al (2012) H5N1 virus causes significant perturbations in host proteome very early in influenza virus-infected primary human monocyte-derived macrophages. J Infect Dis 206:640–645. https://doi.org/10.1093/infdis/jis423
Chou YC, Lai MM, Wu YC, Hsu NC, Jeng KS, Su WC (2015) Variations in genome-wide RNAi screens: lessons from influenza research. J Clin Bioinf 5:2. https://doi.org/10.1186/s13336-015-0017-5
De Santo C et al (2008) Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest 118:4036–4048. https://doi.org/10.1172/JCI36264
Dhillon IS, Sra S (2005) Generalized nonnegative matrix approximations with Bregman divergences. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.861.820&rep=rep1&type=pdf
Dierkes R, Warnking K, Liedmann S, Seyer R, Ludwig S, Ehrhardt C (2014) The Rac1 inhibitor NSC23766 exerts anti-influenza virus properties by affecting the viral polymerase complex activity. PloS One 9:e88520. https://doi.org/10.1371/journal.pone.0088520
Dufner A, Thomas G (1999) Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res 253:100–109. https://doi.org/10.1006/excr.1999.4683
Ehrhardt C et al (2007) Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J Virol 81:3058–3067. https://doi.org/10.1128/Jvi.02082-06
Fan N, Wang J (2016) MicroRNA 34a contributes to virus-mediated apoptosis through binding to its target gene Bax in influenza A virus infection. Biomed Pharmacother 83:1464–1470. https://doi.org/10.1016/j.biopha.2016.08.049
Fan Y et al (2017) Cell cycle-independent role of cyclin D3 in host restriction of influenza virus infection. J Biol Chem 292:5070–5088. https://doi.org/10.1074/jbc.M117.776112
Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, Haagmans BL, McMillan L, Miller DR, Rosenzweig E, Valdar W, Wang J, Churchill GA, Threadgill DW, McWeeney SK, Katze MG, Pardo-Manuel de Villena F, Baric RS, Heise MT (2013) Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathogens 9:e1003196. https://doi.org/10.1371/journal.ppat.1003196
Fujioka Y, Tsuda M, Hattori T, Sasaki J, Sasaki T, Miyazaki T, Ohba Y (2011) The Ras-PI3K signaling pathway is involved in clathrin-independent endocytosis and the internalization of influenza viruses. PloS One 6:e16324. https://doi.org/10.1371/journal.pone.0016324
Fujioka Y et al (2013) A Ca(2+)-dependent signalling circuit regulates influenza A virus internalization and infection. Nat Commun 4:2763. https://doi.org/10.1038/ncomms3763
Gaur P, Munjhal A, Lal SK (2011) Influenza virus and cell signaling pathways. Med Sci Monit 17:RA148–RA154
Geiss GK, An MC, Bumgarner RE, Hammersmark E, Cunningham D, Katze MG (2001) Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J Virol 75:4321–4331. https://doi.org/10.1128/Jvi.75.9.4321-4331.2001
Han J et al (2013) Clinical presentation and sequence analyses of HA and NA antigens of the novel H7N9 viruses. Emerg Microbes Infect 2:e23. https://doi.org/10.1038/emi.2013.28
Hao LH et al (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–U846. https://doi.org/10.1038/nature07151
He Y et al (2010) Influenza A virus replication induces cell cycle arrest in G0/G1 phase. J Virol 84:12832–12840. https://doi.org/10.1128/JVI.01216-10
Heilman DW, Green MR, Teodoro JG (2005) The anaphase promoting complex: a critical target for viral proteins and anti-cancer drugs. Cell Cycle 4:560–563
Jiang W, Wang Q, Chen S, Gao S, Song L, Liu P, Huang W (2013) Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J Virol 87:3039–3052. https://doi.org/10.1128/JVI.03176-12
Karlas A et al (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–U132. https://doi.org/10.1038/nature08760
Kuleshov MV et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. https://doi.org/10.1093/nar/gkw377
Lakadamyali M, Rust MJ, Zhuang X (2004) Endocytosis of influenza viruses Microbes and infection / Institut Pasteur. Microbes Infect 6(10):929–936
Lam TT-Y et al. (2013) The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature advance online publication. doi:https://doi.org/10.1038/nature12515. http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature12515.html#supplementary-information
Lee ASY, Burdeinick-Kerr R, Whelan SPJ (2013) A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA 110:324–329. https://doi.org/10.1073/pnas.1216454109
Li Y, Liang C, Wong KC, Luo J, Zhang Z (2014) Mirsynergy: detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics 30:2627–2635. https://doi.org/10.1093/bioinformatics/btu373
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mainiero F et al (2000) RAC1/P38 MAPK signaling pathway controls beta1 integrin-induced interleukin-8 production in human natural killer cells. Immunity 12:7–16
Nadeau JH (2001) Modifier genes in mice and humans. Nat Rev Genet 2:165–174. https://doi.org/10.1038/35056009
Nedelko T et al (2012) Distinct gene loci control the host response to influenza H1N1 virus infection in a time-dependent manner. BMC Genomics 13:411. https://doi.org/10.1186/1471-2164-13-411
Nencioni L, de Chiara G, Sgarbanti R, Amatore D, Aquilano K, Marcocci ME, Serafino A, Torcia M, Cozzolino F, Ciriolo MR, Garaci E, Palamara AT (2009) Bcl-2 expression and p38MAPK activity in cells infected with influenza A virus: impact on virally induced apoptosis and viral replication. J Biol Chem 284:16004–16015. https://doi.org/10.1074/jbc.M900146200
Nencioni L et al (2003) Influenza A virus replication is dependent on an antioxidant pathway that involves GSH and Bcl-2. FASEB J 17:758–760. https://doi.org/10.1096/fj.02-0508fje
Olsen CW, Kehren JC, Dybdahl-Sissoko NR, Hinshaw VS (1996) bcl-2 alters influenza virus yield, spread, and hemagglutinin glycosylation. J Virol 70:663–666
Parnell G, McLean A, Booth D, Huang S, Nalos M, Tang B (2011) Aberrant cell cycle and apoptotic changes characterise severe influenza A infection—a meta-analysis of genomic signatures in circulating leukocytes. PloS One 6:e17186. https://doi.org/10.1371/journal.pone.0017186
Pommerenke C, Wilk E, Srivastava B, Schulze A, Novoselova N, Geffers R, Schughart K (2012) Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses. PloS One 7:e41169. https://doi.org/10.1371/journal.pone.0041169
Riccadonna S, Jurman G, Visintainer R, Filosi M, Furlanello C (2016) DTW-MIC coexpression networks from time-course data. PloS One 11:e0152648. https://doi.org/10.1371/journal.pone.0152648
Ryan-Poirier KA, Kawaoka Y (1993) Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology 193:974–976. https://doi.org/10.1006/viro.1993.1208
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Shapira SD et al (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. https://doi.org/10.1016/j.cell.2009.12.018
Sieczkarski SB, Whittaker GR (2003) Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333–343
Srivastava B, Błażejewska P, Heßmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K (2009) Host genetic background strongly influences the response to influenza a virus infections. PloS One 4:e4857. https://doi.org/10.1371/journal.pone.0004857
Staring J et al (2017) PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541:412-+. https://doi.org/10.1038/nature21032
Szklarczyk D et al (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. https://doi.org/10.1093/nar/gku1003
Tandon R, Sra S (2010) Sparse nonnegative matrix approximation: new formulations and algorithms. http://www.cyberneum.de/fileadmin/user_upload/files/publications/MPIK-TR-193_[0].pdf
Toth LA, Williams RW (1999) A quantitative genetic analysis of slow-wave sleep in influenza-infected CXB recombinant inbred mice. Behav Genet 29:339–348
Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K (2000) RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 20:2066–2074. https://doi.org/10.1128/Mcb.20.6.2066-2074.2000
Walsh D, Mohr I (2011) Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9:860–875. https://doi.org/10.1038/nrmicro2655
Wang ZF et al (2014) Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci USA 111:769–774. https://doi.org/10.1073/pnas.1321748111
Xiong X et al (2013) Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. https://doi.org/10.1038/nature12372 http://www.nature.com/nature/journal/v499/n7459/abs/nature12372.html#supplementary-information
Yeung M-L et al (2016) MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nature Microbiol 1:16004. https://doi.org/10.1038/nmicrobiol.2016.4 http://www.nature.com/articles/nmicrobiol20164#supplementary-information
Yu L, Sun L, Nan Y, Zhu LY (2011) Protection from H1N1 influenza virus infections in mice by supplementation with selenium: a comparison with selenium-deficient mice. Biol Trace Elem Res 141:254–261. https://doi.org/10.1007/s12011-010-8726-x
Zhao XF et al (2014) PI3K/Akt signaling pathway modulates influenza virus induced mouse alveolar macrophage polarization to M1/M2b. PloS One 9:e104506. https://doi.org/10.1371/journal.pone.0104506
Zhou J et al. (2013) Biological features of novel avian influenza A (H7N9) virus. Nature advance online publication doi:https://doi.org/10.1038/nature12379. http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature12379.html#supplementary-information
Zhu H et al (2013) Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. https://doi.org/10.1126/science.1239844
Funding
This work was supported by grants from the Health and Medical Research Fund of Hong Kong Government (01121726, RRG-08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.Q.S, M.L.Y., K.Y.Y., K.K.W.T., S.H., and S.B. designed research; L.J., Z.G.Z., H.W.L.W., Z.Y., NG., and D.SM.W. performed experiments; S.B., X.Z, and Y.F. analyzed results; and Y.Q.S, M.L.Y., S.B., and X.Z wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Committee of the Use of Live Animals in Teaching and Research (CULATR 3275-14).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Additional File 1
Supplementary figures. Supplementary Fig. S1 showed error bars of virus load in each strain at each day, with mean values and standard deviations calculated from the virus load of all biological replicates at the same day. Supplementary Fig. S2 showed the differential expression levels (log2-fold change) of 10 interferon-stimulated genes between infected and mock-treated mice at day 3. All transcripts for the same gene were presented. Different strains were represented by different colours as shown in the legend. Supplementary Fig. S3 showed the characteristics of genes differentially expressed between C57BL/6J and DBA/2J over time. (A) Heatmap of genes differentially expressed between strains (strain-specific DEGs) showed distinct expression patterns between C57BL/6J and DBA/2J mice. (B) PCA of all samples based on normalized counts of strain-specific DEGs. PC2 revealed distinct transcriptome profile of these DEGs for the two strains due to their genetic background explaining around 26% of the expression variation. Supplementary Fig. S4 demonstrated that genes responsible for host resistance were enriched in DTW-MIC similarities between 0.2-0.25. (A) Histogram of DTW-MIC similarity scores showed two similarity peaks as: between 0.2-0.25 and between 0.7-0.75. (B) PCA of all samples based on normalized counts of genes within different DTW-MIC similarity ranges. The expression variation across samples based on genes with similarity scores between 0.2-0.25 mainly explained the difference from host genetic background, whereas the expression variation based on genes with similarity scores higher than 0.25 were mainly attributed to host response to influenza infection. Supplementary Fig. S5-S6 showed the temporal dynamics of representative genes enriched in cell cycle and ribosome in C57BL/6J and DBA/2J. Supplementary Fig. S7 demonstrated the differential characteristics of time course strain-specific DEGs with high and low temporal similarities between C57BL/6J and DBA/2J. (A) DTW-MIC distribution of all strain-specific DEGs over time. (B) PCA of all samples based on the transcription profiles of strain-specific DEGs with DTW-MIC scores lower than 0.25 (left) and higher than 0.7 (right) respectively C). The same transcription profiles used in B), but presented as heatmaps (DOC 2565 kb)
Additional File 2
Supplementary Tables. Supplementary Table S1: Strain-specific DEGs over time. Supplementary Table S2: The DTW-MIC similarity scores and permutation p-values of all genes. Supplementary Table S3: Summary statistics of DTW-MIC similarities. Supplementary Table S4: Host factors responsible for influenza virus replication show significantly differential DTW-MIC similarities between C57BL/6J and DBA/2J. Supplementary Table S5: Host factors interacting with influenza virus show significantly differential DTW-MIC similarities between C57BL/6J and DBA/2J. Supplementary Table S6: The distribution of DTW-MIC similarity scores of host factors either responsible for influenza virus replication or interact with virus. Supplementary Table S7: The list of candidate genes with both significant differential expression levels and distinct temporal expression patterns between C57BL/6J and DBA/2J. Supplementary Table S8: Functional enrichment analysis of candidate genes with both significant differential expression level and distinct temporal expression pattern between C57BL/6J and DBA/2J. Supplementary Table S9: The list of candidate genes which show distinct temporal expression patterns between C57BL/6J and DBA/2J but have no significant differential expression levels between the two mouse strains. Supplementary Table S10: Four clusters of sig.low.simi genes according to the differential expression levels between infected and mock-treated mice over time in both C57BL/6J and DBA/2J strains (XLS 3855 kb)
Additional File 3
The main functional modules in the interaction network of sig.low.simi genes from clusters 1, 2 and 3 (XLSX 508 kb)
Additional File 4
The modules in the integrated miRNA regulatory network (XLSX 35 kb)
Rights and permissions
About this article
Cite this article
Bao, S., Jia, L., Zhou, X. et al. Integrated analysis of mRNA-seq and miRNA-seq for host susceptibilities to influenza A (H7N9) infection in inbred mouse lines. Funct Integr Genomics 18, 411–424 (2018). https://doi.org/10.1007/s10142-018-0602-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0602-3