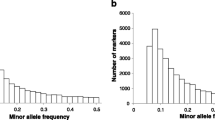
Abstract
Genome selection is mainly used in disease-resistant traits of aquatic species; however, its implementation is hindered by a high cost of genotype and phenotype data collection. Single-step genomic best linear unbiased prediction (SSGBLUP) can integrate phenotypes, genetic markers, and pedigree records into simultaneous prediction without increasing genotyping costs. The objective of this study is to investigate the performance of SSGBLUP in large yellow croaker and to evaluate the effects of the number of phenotypic records and genotyping per family on the predictive ability of SSGBLUP. A large yellow croaker population consists of 6898 individuals from 14 families with survival time resistant against Cryptocaryon irritans (C. irritans), body weight (BW), and body length (BL) traits were collected, of which 669 individuals were genotyped. Results showed that the mean predictive ability of all traits in the individuals randomly sampling for SSGBLUP, GBLUP, and BLUP was 0.738, 0.738, and 0.736, respectively. Moreover, the predictive ability of SSGBLUP and BLUP models did not increase with the extra phenotypic records per family, in which the predictive ability of SSGBLUP and BLUP in survival time was 0.853 and 0.851 for only genotyped data (N = 0) used, and 0.852, 0.845 for all phenotypic records (N = 600) used, respectively. However, with the increase in the genotype number of training set, the prediction ability of SSGBLUP and GBLUP model was increased and the highest predictive ability was gained when the genotype number per family was 40 or 45. In addition, the prediction ability of SSGBLUP model was higher than that of GBLUP. Our study showed that the SSGBLUP model still has great potential and advantages in genomic breeding of large yellow croakers. It is recommended that each family provide 100 phenotypic individuals, of which 40 individuals with genotyping data for SSGBLUP model prediction and family resistance evaluation.


Similar content being viewed by others
Data Availability
The data used to support to the findings of this study are available from the corresponding author upon reasonable request.
References
Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ (2010) Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci 93:743–752
Barria A, Christensen KA, Yoshida GM, Correa K, Jedlicki A, Lhorente JP, Davidson WS, Yanez JM et al (2018) Genomic predictions and genome-wide association study of resistance against piscirickettsia salmonis in coho salmon (Oncorhynchus kisutch) Using ddRAD Sequencing. G3-Genes Genom Genet 8:1183–1194
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Bureau of Fishery Administration of the Ministry of Agriculture and Rural Affairs, N.F.T.E.C (2021) China Fishery Statistical Yearbook 2021. China Society of Fisheries 20–21
Chen Y, Huang W, Shan X, Chen J, Weng H, Yang T, Wang H (2020) Growth characteristics of cage-cultured large yellow croaker Larimichthys crocea. Aquaculture Reports 16:100242
Dagnachew B, Meuwissen T (2019) Accuracy of within-family multi-trait genomic selection models in a sib-based aquaculture breeding scheme. Aquaculture 505:27–33
Dai P, Kong J, Liu J, Lu X, Sui J, Meng X, Luan S et al (2020) Evaluation of the utility of genomic information to improve genetic evaluation of feed efficiency traits of the Pacific white shrimp Litopenaeus vannamei. Aquaculture 527
Garcia ALS, Bosworth B, Waldbieser G, Misztal I, Tsuruta S, Lourenco DAL (2018) Development of genomic predictions for harvest and carcass weight in channel catfish. Genet Sel Evol 50:66
Houston RD, Haley CS, Hamilton A, Guy DR, Tinch AE, Taggart JB, McAndrew BJ, Bishop SC (2008) Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178:1109–1115
Houston RD, Bean TP, Macqueen DJ, Gundappa MK, Jin YH, Jenkins TL, Selly SLC, Martin SAM, Stevens JR, Santos EM, Davie A, Robledo D (2020) Harnessing genomics to fast-track genetic improvement in aquaculture. Nat Rev Genet 21:389–409
Legarra A, Aguilar I, Misztal I (2009) A relationship matrix including full pedigree and genomic information. J Dairy Sci 92:4656–4663
Li X, Zhang Z, Liu X, Chen Y (2019) Impact of genotyping strategy on the accuracy of genomic prediction in simulated populations of purebred swine. Animal 13:1804–1810
Li Y, Jiang B, Mo Z, Li A, Dan X (2022) Cryptocaryon irritans (Brown, 1951) is a serious threat to aquaculture of marine fish. Rev Aquac 14:218–236
Misztal I, Lourenco D, Legarra A (2020) Current status of genomic evaluation. J Anim Sci 98(4)
Norris A (2017) Application of genomics in salmon aquaculture breeding programs by Ashie Norris who knows where the genomic revolution will lead us? Mar Genomics 36:13–15
Pante MJR, Gjerde B, McMillan I, Misztal I (2002) Estimation of additive and dominance genetic variances for body weight at harvest in rainbow trout, Oncorhynchus mykiss. Aquaculture 204:383–392
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Sae-Lim P, Kause A, Lillehammer M, Mulder HA (2017) Estimation of breeding values for uniformity of growth in Atlantic salmon (Salmo salar) using pedigree relationships or single-step genomic evaluation. Genet Sel Evol 49:33
Song H, Dong T, Hu M, Yan X, Xu S, Hu H et al (2022) First single-step genomic prediction and genome-wide association for body weight in Russian sturgeon (Acipenser gueldenstaedtii). Aquaculture 561:738–713
Vallejo RL, Leeds TD, Fragomeni BO, Gao G, Hernandez AG, Misztal I, Welch TJ, Wiens GD, Palti Y (2016) Evaluation of genome-enabled selection for bacterial cold water disease resistance using progeny performance data in rainbow trout: insights on genotyping methods and genomic prediction models. Front Genet 7:96
Vallejo RL, Leeds TD, Gao G, Parsons JE, Martin KE, Evenhuis JP, Fragomeni BO, Wiens GD, Palti Y (2017) Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet Sel Evol 49:17
Wang J, Zhao J, Tong B, Ke Q, Bai Y, Gong J, Zeng J, Deng Y, Lan B, Zhou T, Xu P (2022) Effects of artificial mating on genomic selection of resistance against Cryptocaryon irritans in large yellow croaker. Aquaculture 561:738617
Wang J, Miao L, Chen B, Zhao J, Ke Q, Pu F, Zhou T, Xu P et al (2023) Development and evaluation of liquid SNP array for large yellow croaker (Larimichthys crocea). Aquaculture 563:739–021
Wientjes YC, Veerkamp RF, Calus MP (2013) The effect of linkage disequilibrium and family relationships on the reliability of genomic prediction. Genetics 193:621–631
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42:565–569
Yin L, Zhang H, Tang Z, Yin D, Fu Y, Yuan X, Li X, Liu X, Zhao S et al (2023) HIBLUP: an integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res 51(8):3501–3512
Zhao J, Zhou T, Bai H, Ke Q, Li B, Bai M, Zhou Z, Pu F, Zheng W, Xu P (2021a) Genome-wide association analysis reveals the genetic architecture of parasite (Cryptocaryon irritans) resistance in large yellow croaker (Larimichthys crocea). Mar Biotechnol 23:242–254
Zhao J, Bai H, Ke Q, Li B, Zhou Z, Wang H, Chen B, Pu F, Zhou T, Xu P et al (2021b) Genomic selection for parasitic ciliate Cryptocaryon irritans resistance in large yellow croaker. Aquaculture 531:735–786
Zhou J, Lin Q, Shao B, Ren D, Li J, Zhang Z, Zhang H (2022) Evaluating the application effect of single-step genomic selection in pig populations. Scientia Agricultura Sinica 55:3042–3049
Funding
This work was supported by the National Key R&D Program of China (2022YFD2401002), the National Science Fund for Distinguished Young Scholars (32225049), the National Natural Science Foundation of China (NSFC) (U21A20264).
Author information
Authors and Affiliations
Contributions
Peng Xu and JiaYing Wang conceived and designed the study. JiaYing Wang, YuLin Bai, XiaoQing Zou, ChengYu Li, JunYi Yang, Ji Zhao, QiaoZhen Ke helped on challenge test and phenotype collection. JiaYing Wang analyzed the data and drafted the manuscript. Peng Xu and Tao Zhou revised the manuscript. All authors have read and validated the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This study was approved by the Animal Care and Use Committee at the College of Ocean and Earth Sciences, Xiamen University. Methods were carried out in accordance with approved guidelines.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Bai, Y., Zou, X. et al. First Genomic Prediction of Single-Step Models in Large Yellow Croaker. Mar Biotechnol 25, 603–611 (2023). https://doi.org/10.1007/s10126-023-10229-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-023-10229-0