Abstract
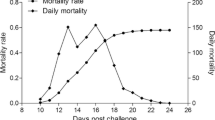
Viral nervous necrosis (VNN) disease caused by the nervous necrosis virus (NNV) is a major disease, leading to a huge economic loss in aquaculture. Previous GWAS and QTL mapping have identified a major QTL for NNV resistance in linkage group 20 in Asian seabass. However, no causative gene for NNV resistance has been identified. In this study, RNA-seq from brains of Asian seabass fingerlings challenged with NNV at four time points (5, 10, 15 and 20 days post-challenge) identified 1228, 245, 189 and 134 DEGs, respectively. Eight DEGs, including rrm1, were located in the major QTL for NNV resistance. An association study in 445 survived and 608 dead fingerlings after NNV challenge revealed that the SNP in rrm1 were significantly associated with NNV resistance. Therefore, rrm1 was selected for functional analysis, as a candidate gene for NNV resistance. The expression of rrm1 was significantly increased in the gill, liver, spleen and muscle, and was suppressed in the brain, gut and skin after NNV challenge. The rrm1 protein was localized in the nuclear membrane. Over-expression of rrm1 significantly decreased viral RNA and titer in NNV-infected Asian seabass cells, whereas knock-down of rrm1 significantly increased viral RNA and titer in NNV-infected Asian seabass cells. The rrm1 knockout heterozygous zebrafish was more susceptible to NNV infection. Our study suggests that rrm1 is one of the causative genes for NNV resistance and the SNP in the gene may be applied for accelerating genetic improvement for NNV resistance.






Similar content being viewed by others
Data Availability
Raw sequence data for the study has been deposited into the DDBJ Sequence Read Archive database with Bioproject no. PRJDB11816.
References
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Angsujinda K, Plaimas K, Smith DR, Kettratad J, Assavalapsakul W (2020) Transcriptomic analysis of red-spotted grouper nervous necrosis virus infected Asian seabass Lates calcarifer (Bloch, 1790). Aqua Rep 18:100517
Chatterjee S, Pal JK (2009) Role of 5′-and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 101:251–262
Chaves-Pozo E, Bandín I, Olveira JG, Esteve-Codina A, Gómez-Garrido J, Dabad M, Alioto T, Esteban MÁ, Cuesta A (2019) European sea bass brain DLB-1 cell line is susceptible to nodavirus: A transcriptomic study. Fish Shellfish Immunol 86:14–24
Chi S, Lo C-F, Kou G, Chang P, Peng S, Chen S (1997) Mass mortalities associated with viral nervous necrosis (VNN) disease in two species of hatchery-reared grouper, Epinephelus fuscogutatus and Epinephelus akaara (Temminck & Schlegel). J Fish Dis 20:185–193
Chong S, Ngoh G, Ng M, Chu K (1987) Growth of lymphocystis virus in a sea bass, Lates calcarifer (Bloch) cell line. Sing Vet J 11:78–85
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Consortium, G.O (2001) Creating the gene ontology resource: design and implementation. Genome Res 11:1425–1433
Danilova N, Bibikova E, Covey TM, Nathanson D, Dimitrova E, Konto Y, Lindgren A, Glader B, Radu CG, Sakamoto KM (2014) The role of the DNA damage response in zebrafish and cellular models of Diamond Blackfan anemia. Dis Mod Mechan 7:895–905
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Engström Y, Rozell B (1988) Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J 7:1615–1620
Food and Agriculture Organization of the United Nations (FAO) (2020) The State of World Fisheries and Aquaculture 2020. Sustainability in action. Rome
Houston RD (2017) Future directions in breeding for disease resistance in aquaculture species. Rev Bras Zoo 46:545–551
Jeong H, Mason SP, Barabási A-L, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411:41–42
Kim J-O, Kim J-O, Kim W-S, Oh M-J (2017) Characterization of the transcriptome and gene expression of brain tissue in sevenband grouper (Hyporthodus septemfasciatus) in response to NNV infection. Genes 8:31
Kümmerer K (2009) Antibiotics in the aquatic environment–a review–part I. Chemosphere 75:417–434
Lamarre S, Frasse P, Zouine M, Labourdette D, Sainderichin E, Hu G, Berre-Anton L, Bouzayen M, Maza E (2018) Optimization of an RNA-Seq differential gene expression analysis depending on biological replicate number and library size. Frontiers Plant Sci 9:108
Leppek K, Das R, Barna M (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19:158
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Liao B-Y, Zhang J (2008) Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci USA 105:6987–6992
Liu P, Wang L, Wan ZY, Ye BQ, Huang S, Wong S-M, Yue GH (2016a) Mapping QTL for resistance against viral nervous necrosis disease in Asian seabass. Mar Biotechnol 18:107–116
Liu P, Wang L, Wong S-M, Yue GH (2016b) Fine mapping QTL for resistance to VNN disease using a high-density linkage map in Asian seabass. Sci Rep 6:32122
Liu P, Wang L, Ye BQ, Huang S, Wong S-M, Yue GH (2017) Characterization of a novel disease resistance gene rtp3 and its association with VNN disease resistance in Asian seabass. Fish Shellfish Immunol 61:61–67
Liu P, (2016) Mapping QTL for resistance to VNN disease and analysis of transcriptome in response to NNV infection in Asian seabass. https://scholarbank.nus.edu.sg/handle/10635/126082. Accessed 25 Aug 2021
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Mai W, Liu H, Chen H, Zhou Y, Chen Y (2018) RGNNV-induced cell cycle arrest at G1/S phase enhanced viral replication via p53-dependent pathway in GS cells. Virus Res 256:142–152
Miller JE, Shivakumar MK, Risacher SL, Saykin AJ, Lee S, Nho K, Kim D, Initiative ASDN (2018) Codon bias among synonymous rare variants is associated with Alzheimer’s disease imaging biomarker. NIH Public Access, Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing
Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S (2007) A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 39:529–533
Munday B, Kwang J, Moody N (2002) Betanodavirus infections of teleost fish: a review. J Fish Dis 25:127–142
Pasaniuc B, Rohland N, Mclaren PJ, Garimella K, Zaitlen N, Li H, Gupta N, Neale BM, Daly MJ, Sklar P (2012) Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet 44:631–635
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619
Pettersen J, Rich K, Jensen BB, Aunsmo A (2015) The economic benefits of disease triggered early harvest: a case study of pancreas disease in farmed Atlantic salmon from Norway. Prevent Vet Med 121:314–324
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497
Rise ML, Hall JR, Rise M, Hori TS, Browne MJ, Gamperl AK, Hubert S, Kimball J, Bowman S, Johnson SC (2010) Impact of asymptomatic nodavirus carrier state and intraperitoneal viral mimic injection on brain transcript expression in Atlantic cod (Gadus morhua). Physiol Genomics 42:266–280
Sagawa M, Ohguchi H, Harada T, Samur MK, Tai Y-T, Munshi NC, Kizaki M, Hideshima T, Anderson KC (2017) Ribonucleotide reductase catalytic subunit M1 (RRM1) as a novel therapeutic target in multiple myeloma. Clin Cancer Res 23:5225–5237
Shen Y, Yue G (2019) Current status of research on aquaculture genetics and genomics-information from ISGA 2018. Aqua Fish 4:43–47
Shen Y, Ma K, Yue GH (2020) Status, challenges and trends of aquaculture in Singapore. Aquaculture 533:736210
Shu Z, Li Z, Huang H, Chen Y, Fan J, Yu L, Wu Z, Tian L, Qi Q, Peng S (2020) Cell-cycle-dependent phosphorylation of RRM1 ensures efficient DNA replication and regulates cancer vulnerability to ATR inhibition. Oncogene 39:5721–5733
Sikorska M, Brewer LM, Youdale T, Richards R, Whitfield JF, Houghten RA, Walker PR (1990) Evidence that mammalian ribonucleotide reductase is a nuclear membrane associated glycoprotein. Biochem Cell Biol 68:880–888
Vij S, Kuhl H, Kuznetsova IS, Komissarov A, Yurchenko AA, Van Heusden P, Singh S, Thevasagayam NM, Prakki SRS, Purushothaman K (2016) Chromosomal-level assembly of the Asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet 12:e1005954
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wang L, Liu P, Huang S, Ye B, Chua E, Wan ZY, Yue GH (2017) Genome-wide association study identifies loci associated with resistance to viral nervous necrosis disease in Asian seabass. Mar Biotechnol 19:255–265
Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875
Yang Z, Wong SM, Yue GH (2020) Characterization of GAB3 and its association with NNV resistance in the Asian seabass. Fish Shellfish Immunol 104:18–24
Yang Z, Yue GH, Wong S-M (2021) VNN disease and status of breeding for resistance to NNV in aquaculture. Aqua Fish, in Press: https://doi.org/10.1016/j.aaf.2021.04.001
Yue GH (2014) Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish 15:376–396
Yue GH, Orban L (2005) A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis 26:3081–3083
Yue G, Orban L, Lim H (2017) Current status of the Asian seabass breeding program. Aquaculture 472:85–85
Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G (2007) DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. New Engl J Med 356:800–808
Acknowledgements
We thank our lab members Mr Hongyan Pang and Yanfei Wen for assistance in spawning, egg collection, larval and juvenile culture. Zituo Yang is supported by a graduate research scholarship from Tropical Marine Science Institute, National University of Singapore
Funding
This research was supported by the internal fund of Temasek Life Sciences Laboratory, Singapore.
Author information
Authors and Affiliations
Contributions
This manuscript is a part of the PhD thesis of ZTY. GHY initiated the selective breeding program of the Asian seabass in 2003. ZTY, SMW and GHY designed the study. ZTY cultured fish, conducted challenge experiments and collected samples. ZTY constructed all experiments, analyzed the data and drafted the manuscript. SMW and GHY supervised the PhD thesis of ZTY. All authors read and approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Z., Wong, S.M. & Yue, G.H. Effects of rrm1 on NNV Resistance Revealed by RNA-seq and Gene Editing. Mar Biotechnol 23, 854–869 (2021). https://doi.org/10.1007/s10126-021-10068-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-021-10068-x