Abstract
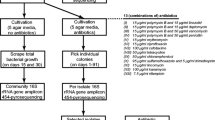
Among 1,236 colony-forming units (CFU) associated with 11 species of marine sponges collected from a Brazilian coast, a total of 100 morphologically different bacterial strains were analyzed. The phylogenetic diversity of the bacterial isolates was assessed by 16S rRNA gene amplification—restriction fragment length polymorphism (RFLP) analysis, using AluI restriction endonuclease. The RFLP fingerprinting resulted in 21 different patterns with good resolution for the identification of the bacterial isolates at the genus level. The genus Bacillus was the most commonly encountered genus, followed by Kocuria. Regarding the relationship between the morphotypes and species of marine sponges, Mycale microsigmatosa presented major diversity, followed by Dragmacidon reticulatum and Polymastia janeirensis. An antibiotic susceptibility profile of the 100 sponge-associated bacterial strains was determined by the disk diffusion method, and we observed a variable resistance profile, with 15 % of the bacteria being multiresistant. In addition, 71 of 100 strains were able to produce biofilm. These 71 strains were divided into 20 strong biofilm producers, 10 moderate biofilm producers, and 41 weak biofilm producers. The plasmid profile of the 100 bacterial strains was analyzed and 38 (38 %) of these samples possessed one or more plasmids. Studies like this are important to increase the information on these associated bacteria found off the coastline of Brazil, a place which has rich biodiversity that is still unknown.


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barros EM, Iório NL, Bastos MC, dos Santos KR, Giambiagi-deMarval M (2007) Species-level identification of clinical staphylococcal isolates based on polymerase chain reaction-restriction fragment length polymorphism analysis of a partial groEL gene sequence. Diagn Microbiol Infect Dis 59(3):251–257
Cetecioğlu Z, Ince BK, Kolukirik M, Ince O (2009) Biogeographical distribution and diversity of bacterial and archaeal communities within highly polluted anoxic marine sediments from the Marmara Sea. Mar Pollut Bull 58(3):384–395
Clinical and Laboratory Standards Institute (CLSI) (2009) Performance standards for antimicrobial susceptibility testing, 19th informational supplement M100-S19. CLSI, Wayne
Davey ME, O’toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64(4):847–867
Delissalde F, Amábile-Cuevas CF (2004) Comparison of antibiotic susceptibility and plasmid content, between biofilm producing and non-producing clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents 24(4):405–408
Fieseler L, Hentschel U, Grozdanov L, Schirmer A, Wen G, Platzer M, Hrvatin S, Butzke D, Zimmermann K, Piel J (2007) Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl Environ Microbiol 73(7):2144–2155
Haddadin RN, Saleh SA, Mahmoud RA, Shehabi AA (2010) Multiple drug resistance and strength of attachment to surfaces in Pseudomonas aeruginosa isolates. Lett Appl Microbiol 51(1):48–53
Hardoim C, Costa R, Araújo F, Hadju E, Peixoto RS, Lins U, Rosado AS, Elsas JDV (2009) Microbial diversity in the marine sponge Aplysina fulva in Brazilian coastal waters. Appl Environ Microbiol 75:3331–3343
Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55(2):167–177
Hooper NJA, van Soest RWM (2002) System Porifera: a guide to the 708 classification of Sponges. Kluwer Academic/Plenum Publishers, New York
Jackson SA, Kennedy J, Morrissey JP, O’Gara F, Dobson ADW (2012) Pyrosequencing reveals diverse and distinct sponge-specific microbial communities in sponges from a single geographical location in Irish waters. Microb Ecol 64(1):105–116
Kim TK, Fuerst JA (2006) Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ Microbiol 8(8):1460–1470
Laport MS, Santos OCS, Muricy G (2009) Marine sponges: potential sources of new antimicrobial drugs. Curr Pharm Biothecnol 10:86–105
Li Z, Liming H, Miao X (2007) Cultivable bacterial community from South China Sea Sponge as revealed by DGGE fingerprinting and 16S rDNA phylogenetic analysis. Curr Microbiol 55:465–472
Marinho PR, Moreira APB, Pellegrino FLPC, Muricy G, Bastos MCF, Dos Santos KRN, Giambiagi-deMarval M, Laport MS (2009) Marine Pseudomonas putida: a potential source of antimicrobial substances against antibiotic-resistant bacteria. Mem Inst Oswaldo Cruz 104:678–682
Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, Egan S, Steinberg P, Kjelleberg S (2008) Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE 3:e2744
Muricy G, Hajdu E (2006) Porifera Brasilis: Guia de Identificação das Esponjas Mais Comuns do Sudeste do Brasil. Eclesiarte, Rio de Janeiro
Phelan RW, Clarke C, Morrissey JP, Dobson AD, O’Gara F, Barbosa TM (2011) Tetracycline resistance-encoding plasmid from Bacillus sp. strain #24, isolated from the marine sponge Haliclona simulans. Appl Environ Microbiol 77(1):327–329
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156
Roberts MC (2011) Environmental macrolide—lincosamide—streptogramin and tetracycline resistant bacteria. Front Microbiol 40(2):1–8
Saá Ibusquiza P, Herrera JJ, Cabo ML (2011) Resistance to benzalkonium chloride, peracetic acid and nisin during formation of mature biofilms by Listeria monocytogenes. Food Microbiol 28(3):418–425
Santos OCS, Pontes PVML, Santos JFM, Muricy G, Giambiagi-Demarval M, Laport MS (2010) Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res Microbiol 161:604–612
Stepanovic S, Vukovic D, Hola V, Bonaventura G, Djukic S, Irkovic I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28(10):2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Turque AS, Cardoso AM, Silveira CB, Vieira RP, Freitas FAD, Albano RM, Gonzalez AM, Paranhos R, Muricy G, Martins OB (2008) Bacterial communities of the marine sponges Hymeniacidon heliophila and Polymastia janeirensis and their environment in Rio de Janeiro, Brazil. Mar Biol 155:135–146
Wang G (2006) Diversity and biotechnological potential of the sponge-associated microbial consortia. J Ind Microbiol Biotechnol 33:545–551
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wilkinson CR, Nowak M, Austin B, Colwell RR (1981) Specificity of bacterial symbionts in Mediterranean and Great Barrier Reef sponge. Microb Ecol 7:13–21
Wright GD (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nature Rev Microbiol 5:175–186
Zhou A, Jiang X, Xu X (1997) Improved alkaline lysis method for rapid isolation of plasmid DNA. Biotechniques 23(4):592–594
Acknowledgments
The authors give special thanks to Dr. Walter Oelemann for his assistance in the preparation of this manuscript. This work was supported by a grant from the CAPES, CNPq, and FAPERJ to M.S. Laport. J. F. Santos-Gandelman is the recipient of a CAPES fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 403 kb)
Rights and permissions
About this article
Cite this article
Santos-Gandelman, J.F., Santos, O.C.S., Pontes, P.V.M. et al. Characterization of Cultivable Bacteria from Brazilian Sponges. Mar Biotechnol 15, 668–676 (2013). https://doi.org/10.1007/s10126-013-9518-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-013-9518-z