Abstract
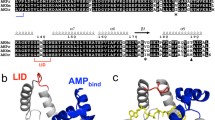
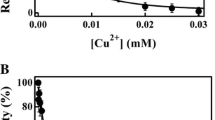
The heterodont clam Calyptogena kaikoi, which inhabits depths exceeding 3,500 m where low ambient temperatures prevail, has an unusual two-domain arginine kinase (AK) with molecular mass of 80 kDa, twice that of typical AKs. The purpose of this work is to investigate the nature of the adaptations of this AK for functioning at low temperatures. Recombinant C. kaikoi AK constructs were expressed, and their two-substrate kinetic constants (k cat, K a, and K ia) were determined at 10°C and 25°C, respectively. When measured at 25°C, the K ia values were tenfold larger than those for corresponding K a values, while at 10°C, the K ia values decreased remarkably, but the K a values were almost unchanged. The Calyptogena two-domain enzyme has threefold higher catalytic efficiency, calculated by k cat/(K ARGa ·K ATPia ), at 10°C, than that at 25°C, reflecting adaptation for function at reduced ambient temperatures. The activation energy (E a) and thermodynamic parameters were determined for Calyptogena two-domain enzyme and compared with those of two-domain enzymes from mesophilic Corbicula and Anthopleura. The value for E a of Calyptogena enzyme were about half of those for mesophilic enzymes, and a larger decrease in entropy was observed in Calyptogena AK reaction. Although large decrease in entropy increases the ΔG o‡ value and consequently lowers the k cat value, this is compensated with its lower E a value thereby minimizing the reduction in its k cat value. These thermodynamic properties, together with the kinetic ones, are also present in the separated domain 2 of the Calyptogena two-domain enzyme.



Similar content being viewed by others
Abbreviations
- AK:
-
Arginine kinase
References
Andrews L, Graham J, Snider M, Fraga D (2008) Characterization of a novel bacterial arginine kinase from Desulfotalea psychrophila. Comp Biochem Physiol B 150:312–319
Arp AJ, Childress JJ (1981) Blood function in the hydrothermal vent vestimentiferan tube worm. Science 213:342–344
Aurilia V, Parracino A, D’Auria S (2008) Microbial carbohydrate esterases in cold adapted environments. Gene 410:234–240
Barman TE, Travers F, Bertrand R, Roseau G (1978) Transient-phase studies on the arginine kinase reaction. Eur J Biochem 89:243–249
Brooks JM, Kennicutt MC 2nd, Fisher CR, Macko SA, Cole K, Childress JJ, Bidigare RR, Vetter RD (1987) Deep-sea hydrocarbon seep communities: evidence for energy and nutritional carbon sources. Science 238:1138–1142
Ciardiello MA, Camardella L, Carratore V, di Prisco G (2000) l-Glutamate dehydrogenase from the antarctic fish Chaenocephalus aceratus. Primary structure, function and thermodynamic characterisation: relationship with cold adaptation. Biochim Biophys Acta 1543:11–23
Cleland WW (1979) Statistical analysis of enzyme kinetic data. Methods Enzymol 63:103–138
Compaan DM, Ellington WR (2003) Functional consequences of a gene duplication and fusion event in an arginine kinase. J Exp Biol 206:1545–1556
Conejo M, Bertin M, Pomponi S, Ellington WR (2008) The early evolution of the phosphagen kinases—insights from choanoflagellate and poriferan arginine kinases. J Mol Evol 66:11–20
Corliss JB, Dymond J, Gordon LI, Edmond JM, von Herzen RP, Ballard RD, Green K, Williams D, Bainbridge A, Crane K, van Andel TH (1979) Submarine thermal sprirngs on the galapagos rift. Science 203:1073–1083
D’Amico S, Gerday C, Feller G (2001) Structural determinants of cold adaptation and stability in a large protein. J Biol Chem 276:25791–25796
D’Amico S, Claverie P, Collins T, Georlette D, Gratia E, Hoyoux A, Meuwis MA, Feller G, Gerday C (2002) Molecular basis of cold adaptation. Philos Trans R Soc Lond B Biol Sci 357:917–925
Edmiston PL, Schavolt KL, Kersteen EA, Moore NR, Borders CL (2001) Creatine kinase: a role for arginine-95 in creatine binding and active site organization. Biochim Biophys Acta 1546:291–298
Ellington WR (1989) Phosphocreatine represents a thermodynamic and functional improvement over other muscle phosphagens. J Exp Biol 143:177–194
Ellington WR (2001) Evolution and physiological roles of phosphagen systems. Annu Rev Physiol 63:289–325
Ellington WR, Suzuki T (2006) Evolution and divergence of creatine kinase genes. In: Vial C (ed) Molecular anatomy and physiology of proteins: creatine kinase. Nova Science, New York, pp 1–27
Feller G, Gerday C (1997) Psychrophilic enzymes: molecular basis of cold adaptation. Cell Mol Life Sci 53:830–841
Fields PA, Houseman DE (2004) Decreases in activation energy and substrate affinity in cold-adapted A4-lactate dehydrogenase: evidence from the Antarctic notothenioid fish Chaenocephalus aceratus. Mol Biol Evol 21:2246–2255
Fields PA, Somero GN (1998) Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci USA 95:11476–11481
Fujimoto N, Tanaka K, Suzuki T (2005) Amino acid residues 62 and 193 play the key role in regulating the synergism of substrate binding in oyster arginine kinase. FEBS Lett 579:1688–1692
Gerday C, Aittaleb M, Arpigny JL, Baise E, Chessa JP, Garsoux G, Petrescu I, Feller G (1997) Psychrophilic enzymes: a thermodynamic challenge. Biochim Biophys Acta 1342:119–131
Grossman SH (1983) Interaction of creatine kinase from monkey brain with substrate: analysis of kinetics and fluorescence polarization. J Neurochem 41:729–736
Hochachka PW, Somero GN (2002) Biochemical adaptation: mechanism and process in physiological evolution. Oxford University, Oxford
Iwanami K, Iseno S, Uda K, Suzuki T (2009) A novel arginine kinase from the shrimp Neocaridina denticulata: the fourth arginine kinase gene lineage. Gene 437:80–87
Joseph B, Ramteke PW, Thomas G (2008) Cold active microbial lipases: some hot issues and recent developments. Biotechnol Adv 26:457–470
Kenyon GL, Reed GH (1983) Creatine kinase: structure-activity relationships. Adv Enzymol Relat Areas Mol Biol 54:367–426
Lonhienne T, Gerday C, Feller G (2000) Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim Biophys Acta 1543:1–10
Lu M, Wang S, Fang Y, Li H, Liu S, Liu H (2010) Cloning, expression, purification, and characterization of cold-adapted alpha-amylase from Pseudoalteromonas arctica GS230. Protein J 29:591–597
Marx JC, Collins T, D’Amico S, Feller G, Gerday C (2007) Cold-adapted enzymes from marine Antarctic microorganisms. Mar Biotechnol (NY) 9:293–304
McLeish M, Kenyon G (2005) Relating structure to mechanism in creatine kinase. Crit Rev Biochem Mol Biol 40:1–20
Morrison JF (1973) Arginine kinase and other invertebrate guanidino kinases. In: Boyer PC (ed) The enzymes. Academic, New York, pp 457–486
Morrison JF, James E (1965) The mechanism of the reaction catalysed by adenosine triphosphate-creatine phosphotransferase. Biochem J 97:37–52
Pereira CA, Alonso GD, Paveto MC, Iribarren A, Cabanas ML, Torres HN, Flawia MM (2000) Trypanosoma cruzi arginine kinase characterization and cloning. A novel energetic pathway in protozoan parasites. J Biol Chem 275:1495–1501
Rao BD, Buttlaire DH, Cohn M (1976) 31P NMR studies of the arginine kinase reaction. Equilibrium constants and exchange rates at stoichiometric enzyme concentration. J Biol Chem 251:6981–6986
Siddiqui KS, Cavicchioli R (2006) Cold-adapted enzymes. Annu Rev Biochem 75:403–433
Simpson BK, Haard NF (1984) Trypsin from Greenland cod, Gadus ogac. Isolation and comparative properties. Comp Biochem Physiol B 79:613–622
Smalas AO, Leiros HK, Os V, Willassen NP (2000) Cold adapted enzymes. Biotechnol Annu Rev 6:1–57
Somero GN (1995) Proteins and temperature. Annu Rev Physiol 57:43–68
Somero GN (2004) Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol 139:321–333
Sotelo-Mundo RR, Lopez-Zavala AA, Garcia-Orozco KD, Arvizu-Flores AA, Velazquez-Contreras EF, Valenzuela-Soto EM, Rojo-Dominguez A, Kanost MR (2007) The lysozyme from insect (Manduca sexta) is a cold-adapted enzyme. Protein Pept Lett 14:774–778
Suzuki T, Kawasaki Y, Furukohri T (1997) Evolution of phosphagen kinase. Isolation, characterization and cDNA-derived amino acid sequence of two-domain arginine kinase from the sea anemone Anthopleura japonicus. Biochem J 328:301–306
Suzuki T, Kawasaki Y, Unemi Y, Nishimura Y, Soga T, Kamidochi M, Yazawa Y, Furukohri T (1998) Gene duplication and fusion have occurred frequently in the evolution of phosphagen kinases—a two-domain arginine kinase from the clam Pseudocardium sachalinensis. Biochim Biophys Acta 1388:253–259
Suzuki T, Kamidochi M, Inoue N, Kawamichi H, Yazawa Y, Furukohri T, Ellington WR (1999) Arginine kinase evolved twice: evidence that echinoderm arginine kinase originated from creatine kinase. Biochem J 340:671–675
Suzuki T, Fukuta H, Nagato H, Umekawa M (2000) Arginine kinase from Nautilus pompilius, a living fossil. Site-directed mutagenesis studies on the role of amino acid residues in the Guanidino specificity region. J Biol Chem 275:23884–23890
Suzuki T, Sugimura N, Taniguchi T, Unemi Y, Murata T, Hayashida M, Yokouchi K, Uda K, Furukohri T (2002) Two-domain arginine kinases from the clams Solen strictus and Corbicula japonica: exceptional amino acid replacement of the functionally important D(62) by G. Int J Biochem Cell Biol 34:1221–1229
Suzuki T, Tomoyuki T, Uda K (2003) Kinetic properties and structural characteristics of an unusual two-domain arginine kinase of the clam Corbicula japonica. FEBS Lett 533:95–98
Tada H, Suzuki T (2010) Cooperativity in the two-domain arginine kinase from the sea anemone Anthopleura japonicus. II. Evidence from site-directed mutagenesis studies. Int J Biol Macromol 47:250–254
Tanaka K, Suzuki T (2004) Role of amino-acid residue 95 in substrate specificity of phosphagen kinases. FEBS Lett 573:78–82
Travers F, Bertrand R, Roseau G, Van Thoai N (1978) Cryoenzymologic studies on arginine kinase: solvent, temperature and pH effects on the overall reaction. Eur J Biochem 88:523–528
Tutino ML, di Prisco G, Marino G, de Pascale D (2009) Cold-adapted esterases and lipases: from fundamentals to application. Protein Pept Lett 16:1172–1180
Uda K, Tanaka K, Bailly X, Zal F, Suzuki T (2005) Phosphagen kinase of the giant tubeworm Riftia pachyptila. Cloning and expression of cytoplasmic and mitochondrial isoforms of taurocyamine kinase. Int J Biol Macromol 37:54–60
Uda K, Fujimoto N, Akiyama Y, Mizuta K, Tanaka K, Ellington WR, Suzuki T (2006) Evolution of the arginine kinase gene family. Comp Biochem Physiol Part D Genom Proteom 1:209–218
Uda K, Yamamoto K, Iwasaki I, Iwai M, Fujikura K, Ellington WR, Suzuki T (2008) Two-domain arginine kinase from the deep-sea clam Calyptogena kaikoi—evidence of two active domains. Comp Biochem Physiol Part B Biochem Mol Biol 151:176–182
Ueda M, Asano T, Nakazawa M, Miyatake K, Inouye K (2008) Purification and characterization of novel raw-starch-digesting and cold-adapted alpha-amylases from Eisenia foetida. Comp Biochem Physiol B Biochem Mol Biol 150:125–130
Ueda M, Goto T, Nakazawa M, Miyatake K, Sakaguchi M, Inouye K (2010) A novel cold-adapted cellulase complex from Eisenia foetida: characterization of a multienzyme complex with carboxymethylcellulase, beta-glucosidase, beta-1,3 glucanase, and beta-xylosidase. Comp Biochem Physiol B Biochem Mol Biol 157:26–32
Watts D, Bannister L (1970) Location of arginine kinase in the cilia of Tetrahymena pyriformis. Nature 226:450–451
Wu X, Ye S, Guo S, Yan W, Bartlam M, Rao Z (2010) Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity. FASEB J 24:242–252
Wyss M, Kaddurah-Daouk R (2000) Creatine and creatinine metabolism. Physiol Rev 80:1107–1213
Wyss M, Smeitink J, Wevers R, Wallimann T (1992) Mitochondrial creatine kinase: a key enzyme of aerobic energy metabolism. Biochim Biophys Acta 1102:119–166
Yousef M, Clark S, Pruett P, Somasundaram T, Ellington W, Chapman M (2003) Induced fit in guanidino kinases—comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci 12:103–111
Zhou G, Somasundaram T, Blanc E, Parthasarathy G, Ellington W, Chapman M (1998) Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions. Proc Natl Acad Sci USA 95:8449–8454
Acknowledgments
We thank W. Ross Ellington, Florida State University, for critically reading the manuscript. This work was supported by a Grant-in-Aid for Scientific Research in Japan to TS (17570062 and 20570072).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suzuki, T., Yamamoto, K., Tada, H. et al. Cold-adapted Features of Arginine Kinase from the Deep-sea Clam Calyptogena kaikoi . Mar Biotechnol 14, 294–303 (2012). https://doi.org/10.1007/s10126-011-9411-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-011-9411-6