Abstract
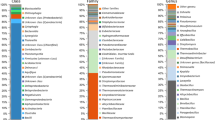
Preliminary characterization of the microbial phylogeny and metabolic potential of a deep-sea sediment sample from the Qiongdongnan Basin, South China Sea, was carried out using a metagenomic library approach. An effective and rapid method of DNA isolation, purification, and library construction was used resulting in approximately 200,000 clones with an average insert size of about 36 kb. End sequencing of 600 individual clones from the fosmid library generated 1,051 sequences with an average sequence length of 619 bp. Phylogenetic ascription indicated that this library was dominated by Bacteria, predominately Proteobacteria, though Planctomycetes were also relatively abundant. Sulfate-reducing and anaerobic ammonium-oxidizing bacteria, which play important roles in the cycling of sedimentary nutrients, were abundant in the library. Cluster of orthologous groups category analysis showed that most of the genes contained in the end sequences were related to metabolism, and with cellular processes and signaling. Functional groups assigned by SEED (subsystems-based annotations) highlighted the existence of ‘one-carbon’ metabolism within this community as well as identifying functional genes involved in methanogenesis. Furthermore, diverse genes involved in the biodegradation of xenobiotics were found using Kyoto Encyclopedia of Genes and Genomes metabolic pathway analysis.





Similar content being viewed by others
References
Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, Villacorta R, Amjadi M, Garrigues C, Jovanovich SB, Feldman RA, DeLong EF (2000) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2:516–529
Biddle J, Fitz-Gibbon S, Schuster S, Brenchley J, House C (2008) Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci U S A 105:10583–10588
Chandler DP, Fredrickson JK, Brockman FJ (1997) Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol 6:475–482
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM (2006) Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311:496–503
Edlund A, Hardeman F, Jansson JK, Sjoling S (2008) Active bacterial community structure along vertical redox gradients in Baltic Sea sediment. Environ Microbiol 10:2051–2063
Ferry JG (1999) Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23:13–38
Fortin N, Beaumier D, Lee K, Greer CW (2004) Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. J Microbiol Methods 56:181–191
Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G (2008) Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol 66:181–196
Guo B, Gu J, Ye Y, Tang Y, Kida K, Wu X (2007) Marinobacter segnicrescens sp. nov., a moderate halophile isolated from benthic sediment of the South China Sea. Int J Syst Evol Microbiol 57:1970–1974
Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF (2004) Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Jiang H, Dong H, Ji S, Ye Y, Wu N (2007) Microbial diversity in the deep marine sediments from the Qiongdongnan Basin in South China Sea. Geomicrobiol J 24:505–517
Kauffmann IM, Schmitt J, Schmid RD (2004) DNA isolation from soil samples for cloning in different hosts. Appl Microbiol Biotechnol 64:665–670
Kennedy J, Marchesi JR, Dobson ADW (2008) Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb Cell Fact 7:27
Lipp JS, Morono Y, Inagaki F, Hinrichs KU (2008) Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454:991–994
Liu Z, Wang B, Liu X, Dai X, Liu Y, Liu S (2008) Paracoccus halophilus sp. nov., isolated from marine sediment of the South China Sea, China, and emended description of genus Paracoccus Davis 1969. Int J Syst Evol Microbiol 58:257–261
Llobet-Brossa E, Rossello-Mora R, Amann R (1998) Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol 64:2691–2696
Ludmann T, Kin Wong H, Wang P (2001) Plio-Quaternary sedimentation processes and neotectonics of the northern continental margin of the South China Sea. Mar Geol 172:331–358
Martin-Cuadrado AB, Lopez-Garcia P, Alba JC, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodriguez-Valera F (2007) Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE 2:e914
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454
Overbeek R, Begley T, Butler R, Choudhuri J, Chuang H, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank E, Gerdes S, Glass E, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy A, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch G, Rodionov D, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702
Parkes RJ, Cragg BA, Wellsbury P (2000) Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol J 8:11–28
Rajendhran J, Gunasekaran P (2008) Strategies for accessing soil metagenome for desired applications. Biotechnol Adv 26:576–590
Riesenfeld CS, Schloss PD, Handelsman J (2004) Metagenomics: genomic analysis of microbial communities. Annu Rev Genet 38:525–552
Roussel EG, Cambon Bonavita MA, Querellou J, Cragg BA, Webster G, Prieur D, Parkes RJ (2008) Extending the deep sub-seafloor biosphere. Science 320:1046
Rusch BD, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC (2007) The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:398–431
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schippers A, Neretin LN (2006) Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ Microbiol 8:1251–1260
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–106
Schneegurt MA, Dore SY, Kulpa CK Jr (2003) Direct extraction of DNA from soils for studies in microbial ecology. Curr Issues Mol Biol 5:1–8
Sowers KR (2004) Methanogenesis. The desk encyclopedia of microbiology. Elsevier Academic, San Diego, pp 659–679
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, Van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten SM (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Su X, Chen F, Yu X, Huang Y (2005) A pilot study on Miocene through Holocene sediments from the continental slope of the South China Sea in correlation with possible distribution of gas hydrates. Geosci 19:1–13
Tao L, Peng W, Pinxian W (2008) Microbial diversity in surface sediments of the Xisha Trough, the South China Sea. Acta Ecol Sinica 28:1166–1173
Tian X, Zhi X, Qiu Y, Zhang Y, Tang S, Xu L, Zhang S, Li W (2009) Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int J Syst Evol Microbiol 59:222–228
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Webster G, Newberry CJ, Fry JC, Weightman AJ (2003) Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J Microbiol Methods 55:155–164
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583
Wu B, Zhang G, Zhu Y, Lu Z, Chen B (2003) Progress of gas hydrate investigation in China offshore. Earth Sci Front 10:177–190
Xu F, Dai X, Chen Y, Zhou H, Cai J, Qu L (2004) Phylogenetic diversity of Bacteria and Archaea in the Nansha marine sediments, as determined by 16S rDNA analysis. Oceanologia et Limnologia Sinica 35:94–96
Ye J, McGinnis S, Madden TL (2006) BLAST: improvements for better sequence analysis. Nucleic Acids Res 34:W6–W9
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zink KG, Wilkes H, Disco U, Elvert M, Horsfield B (2003) Intact phospholipids—microbial “life markers” in marine deep subsurface sediments. Org Geochem 34:755–769
Acknowledgments
This work was partly supported by the National High Technology Research and Development Program of China (2007AA09Z443 and 2007AA021301) and Knowledge Innovation Project of The Chinese Academy of Sciences (KSCX2-YW-G-022).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yongfei Hu and Chengzhang Fu contributed equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material Table 1
(XLS 290 kb)
Supplementary material Table 2
(XLS 162 kb)
Supplementary material Table 3
(XLS 39 kb)
Supplementary material Table 4
(DOC 112 kb)
Rights and permissions
About this article
Cite this article
Hu, Y., Fu, C., Yin, Y. et al. Construction and Preliminary Analysis of a Deep-Sea Sediment Metagenomic Fosmid Library from Qiongdongnan Basin, South China Sea. Mar Biotechnol 12, 719–727 (2010). https://doi.org/10.1007/s10126-010-9259-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-010-9259-1