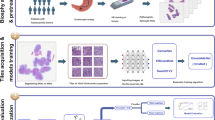
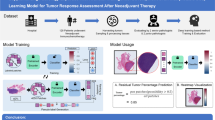
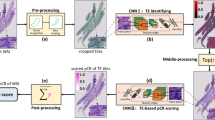
Abstract
Background
Neoadjuvant chemotherapy (NAC) has been recognized as an effective therapeutic option for locally advanced gastric cancer as it is expected to reduce tumor size, increase the resection rate, and improve overall survival. However, for patients who are not responsive to NAC, the best operation timing may be missed together with suffering from side effects. Therefore, it is paramount to differentiate potential respondents from non-respondents. Histopathological images contain rich and complex data that can be exploited to study cancers. We assessed the ability of a novel deep learning (DL)-based biomarker to predict pathological responses from images of hematoxylin and eosin (H&E)-stained tissue.
Methods
In this multicentre observational study, H&E-stained biopsy sections of patients with gastric cancer were collected from four hospitals. All patients underwent NAC followed by gastrectomy. The Becker tumor regression grading (TRG) system was used to evaluate the pathologic chemotherapy response. Based on H&E-stained slides of biopsies, DL methods (Inception-V3, Xception, EfficientNet-B5, and ensemble CRSNet models) were employed to predict the pathological response by scoring the tumor tissue to obtain a histopathological biomarker, the chemotherapy response score (CRS). The predictive performance of the CRSNet was evaluated.
Results
69,564 patches from 230 whole-slide images of 213 patients with gastric cancer were obtained in this study. Based on the F1 score and area under the curve (AUC), an optimal model was finally chosen, named the CRSNet model. Using the ensemble CRSNet model, the response score derived from H&E staining images reached an AUC of 0.936 in the internal test cohort and 0.923 in the external validation cohort for predicting pathological response. The CRS of major responders was significantly higher than that of minor responders in both internal and external test cohorts (both p < 0.001).
Conclusion
In this study, the proposed DL-based biomarker (CRSNet model) derived from histopathological images of the biopsy showed potential as a clinical aid for predicting the response to NAC in patients with locally advanced GC. Therefore, the CRSNet model provides a novel tool for the individualized management of locally advanced gastric cancer.






Similar content being viewed by others
Data availability
Data is available under request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet (London, England). 2020;396(10251):635–48.
Pelc Z, Skórzewska M, Rawicz-Pruszyński K, Polkowski WP. Lymph node involvement in advanced gastric cancer in the era of multimodal treatment-oncological and surgical perspective. Cancers. 2021;13(10):2509.
Gervaso L, Pellicori S, Cella CA, Bagnardi V, Lordick F, Fazio N. Biomarker evaluation in radically resectable locally advanced gastric cancer treated with neoadjuvant chemotherapy: an evidence reappraisal. Ther Adv Med Oncol. 2021;13:17588359211029560.
Sun J, Wang X, Zhang Z, Zeng Z, Ouyang S, Kang W. The sensitivity prediction of neoadjuvant chemotherapy for gastric cancer. Front Oncol. 2021;11: 641304.
Wang W, Peng Y, Feng X, Zhao Y, Seeruttun SR, Zhang J, Cheng Z, Li Y, Liu Z, Zhou Z. Development and validation of a computed tomography-based radiomics signature to predict response to neoadjuvant chemotherapy for locally advanced gastric cancer. JAMA Netw Open. 2021;4(8): e2121143.
Lowy AM, Mansfield PF, Leach SD, Pazdur R, Dumas P, Ajani JA. Response to neoadjuvant chemotherapy best predicts survival after curative resection of gastric cancer. Ann Surg. 1999;229(3):303–8.
Langer R, Becker K. Tumor regression grading of gastrointestinal cancers after neoadjuvant therapy. Virchows Archiv Int J Pathol. 2018;472(2):175–86.
Neves Filho EH, de Sant’Ana RO, Nunes LV, Pires AP, da Cunha MD. Histopathological regression of gastric adenocarcinoma after neoadjuvant therapy: a critical review. APMIS Acta Pathol Microbiol Immunol Scand. 2017;125(2):79–84.
Deng S, Zhang X, Yan W, Chang EI, Fan Y, Lai M, Xu Y. Deep learning in digital pathology image analysis: a survey. Front Med. 2020;14(4):470–87.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500–10.
Bilal M, Raza SEA, Azam A, Graham S, Ilyas M, Cree IA, Snead D, Minhas F, Rajpoot NM. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health. 2021;63:1608.
Farahmand S, Fernandez AI, Ahmed FS, Rimm DL, Chuang JH, Reisenbichler E, Zarringhalam K. Deep learning trained on hematoxylin and eosin tumor region of Interest predicts HER2 status and trastuzumab treatment response in HER2+ breast cancer. Mod Pathol Off J US Can Acade Pathol. 2021;15:2894.
Li F, Yang Y, Wei Y, He P, Chen J, Zheng Z, Bu H. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J Transl Med. 2021;19(1):348.
Hayashi M, Fujita T, Matsushita H. Prognostic value of tumor regression grade following the administration of neoadjuvant chemotherapy as treatment for gastric/gastroesophageal adenocarcinoma: a meta-analysis of 14 published studies. Eur J Surg Oncol J Eur Soc Surg Oncol Br Assoc Surg Oncol. 2021;47(8):1996–2003.
Pouw RE, Barret M, Biermann K, Bisschops R, Czakó L, Gecse KB, de Hertogh G, Hucl T, Iacucci M, Jansen M, et al. Endoscopic tissue sampling—part 1: upper gastrointestinal and hepatopancreatobiliary tracts. European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy. 2021;53(11):1174–88.
Muti HS, Heij LR, Keller G, Kohlruss M, Langer R, Dislich B, Cheong JH, Kim YW, Kim H, Kook MC, et al. Development and validation of deep learning classifiers to detect Epstein-Barr virus and microsatellite instability status in gastric cancer: a retrospective multicentre cohort study. Lancet Digit Health. 2021;3(10):e654–64.
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–67.
Wu Z, Lin Z, Li L, Pan H, Chen G, Fu Y, Qiu Q. Deep learning for classification of pediatric otitis media. Laryngoscope. 2021;131(7):E2344-e2351.
Hou JU, Park SW, Park SM, Park DH, Park CH, Min S. Efficacy of an artificial neural network algorithm based on thick-slab magnetic resonance cholangiopancreatography images for the automated diagnosis of common bile duct stones. J Gastroenterol Hepatol. 2021;36(12):3532–40.
Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122-1131.e1129.
Tong Y, Liu D, Zhang J. Connection and distinction of tumor regression grading systems of gastrointestinal cancer. Pathol Res Pract. 2020;216(9): 153073.
Sato Y, Okamoto K, Kawaguchi T, Nakamura F, Miyamoto H, Takayama T. Treatment response predictors of neoadjuvant therapy for locally advanced gastric cancer: current status and future perspectives. Biomedicines. 2022;10(7):1614.
Chen YH, Xiao J, Chen XJ, Wang HS, Liu D, Xiang J, Peng JS. Nomogram for predicting pathological complete response to neoadjuvant chemotherapy in patients with advanced gastric cancer. World J Gastroenterol. 2020;26(19):2427–39.
Meier A, Nekolla K, Hewitt LC, Earle S, Yoshikawa T, Oshima T, Miyagi Y, Huss R, Schmidt G, Grabsch HI. Hypothesis-free deep survival learning applied to the tumour microenvironment in gastric cancer. J Pathol Clin Res. 2020;6(4):273–82.
Huang B, Tian S, Zhan N, Ma J, Huang Z, Zhang C, Zhang H, Ming F, Liao F, Ji M, et al. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: a retrospective multicentre study. EBioMedicine. 2021;73: 103631.
Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21(11):4012.
Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–15.
Salvi M, Acharya UR, Molinari F, Meiburger KM. The impact of pre- and post-image processing techniques on deep learning frameworks: a comprehensive review for digital pathology image analysis. Comput Biol Med. 2021;128: 104129.
Castiglioni I, Rundo L, Codari M, Di Leo G, Salvatore C, Interlenghi M, Gallivanone F, Cozzi A, D’Amico NC, Sardanelli F. AI applications to medical images: from machine learning to deep learning. Phys Medica PM. 2021;83:9–24.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81602049 and 81802342), the Natural Science Foundation of Guangdong Province, China (Grant No. 2018A030313978), Shenzhen Science and Technology Program (No. JCYJ20220530145001002), and the Kelin New Star of the First Affiliated Hospital of Sun Yat‐Sen University (Grant Nos. R08011 and R08010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no conflict of interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, Z., Ren, Y., Zhang, Z. et al. Digital histopathological images of biopsy predict response to neoadjuvant chemotherapy for locally advanced gastric cancer. Gastric Cancer 26, 734–742 (2023). https://doi.org/10.1007/s10120-023-01407-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-023-01407-z