Abstract
Background
A disintegrin and metalloproteinase with thrombospondin motifs 10 (ADAMTS10) plays a role in extracellular matrix and correlates with Weill–Marchesani syndrome. However, its role in gastric cancer remains unknown. Thus, we started this research to unveil the role of ADAMTS10 in gastric cancer (GC).
Methods
The expression of ADAMTS10 in GC was analyzed by immunohistochemical staining and quantitative RT-PCR (qRT-PCR). The effects of ADAMTS10 inhibiting GC cell progression were conducted by functional experiments in vitro and in vivo. Flow cytometry was used to discover changing of cell cycle, apoptosis and ROS by ADAMTS10 in GC cell. Western blot was applied to identify targets of ADAMTS10. Western blot, qRT-PCR and flow cytometry were applied to discover the effect of ADAMT10 on THP1.
Results
ADAMTS10 expression was downregulated in GC tissue and patients with low ADAMTS10 levels had poorer overall survival. ADAMTS10 overexpression altered cell cycle, promoted apoptosis, and inhibited proliferation, migration, and invasion in vitro and in vivo. ADAMTS10 regulated TXNIP and ROS through the JAK/STAT/c-MYC pathway. Decreasing TXNIP and ROS reversed the inhibitory effect of ADAMTS10 on cell migration and invasion in vitro. ADAMTS10 secreted by GC cells was absorbed by THP1 and regulated TXNIP and ROS in THP1. ADAMTS10 secreted by GC cells inhibited macrophage M2 polarization.
Conclusions
These results suggest that ADAMTS10 targets TXNIP and ROS via the JAK/STAT/c-MYC pathway and that may play important roles in GC progression and macrophage polarization which indicates that ADAMTS10 can be a potential survival marker for gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the third most common cause of cancer-related deaths worldwide, with over 1,000,000 new cases and approximately 784,000 associated deaths in 2018 [1]. Despite advances in its diagnosis and treatment in recent years, the prognosis of GC remains quite poor compared to other solid tumors. This is partly because most patients have already developed advanced disease by the time of diagnosis and partly because the mechanisms of tumorigenesis, growth, and migration are not fully understood, and no treatment can cure all GCs. Therefore, it is crucial to unravel the molecular mechanisms underlying the development and growth of GC and to find metastasis predictors or therapeutic targets to achieve a better prognosis.
The A Disintegrin and Metalloproteinase with Thrombospondin motifs (ADAMTS) family includes 19 members involved in various important biological processes, such as hemostasis, extracellular matrix (ECM) remodeling, and regulation of angiogenesis associated with carcinogenesis and metastasis [2]. Abnormal expression or function of ADAMTS family members is associated with connective tissue disease, inflammation, arthritis, cancer, and thrombotic thrombocytopenia [3,4,5,6,7,8,9]. ADAMTS10 belongs to the secreted protein superfamily and plays a role in the structure and function of microfibrils by interacting with fibrillin-1 and 2 and contributing to ECM formation and conversion [10,11,12]. ADAMTS10 recessive mutations lead to Weill–Marchesani syndrome, which is associated with short stature, skin and cornea thickening, heart valve fibrosis, and lens defects [13,14,15]. However, the role of ADAMTS10 in cancer, particularly in GC, has not been studied.
The JAK/STAT pathway is a highly conserved signal transduction pathway. It regulates a variety of cellular mechanisms associated with the development of multiple diseases. Continuously aberrant activation of the JAK/STAT pathway is associated with malignancies such as leukemia, Hodgkin's lymphoma, myeloproliferative neoplasms, natural killer/T-cell lymphoma, hepatocellular carcinoma, lung cancer, head and neck cancer, and prostate cancer. At present, the research of JAK / STAT pathway in gastric cancer is still insufficient, therefore, it is very promising to study the role and mechanism of JAK / STAT pathway in gastric cancer [16].
Thioredoxin-interacting protein (TXNIP), also known as vitamin D3 upregulated protein 1 (VDUP1) [17] and thioredoxin-binding protein-2 (TBP-2) [18], is known to be a multifunctional protein [19]. TXNIP binds to thioredoxin, inhibiting its antioxidant function [20]. It can also promote insulin secretion, inhibit glucose uptake, and reprogram metabolism [21,22,23]. TXNIP is also considered a tumor suppressor protein due to its effects on tumor cell proliferation, apoptosis, cell cycle, and metastasis [24,25,26]. However, the expression and biological functions of TXNIP in GC are largely unknown.
In this study, we investigated the expression levels of ADAMTS10 in GC tissue and cells and analyzed the relationship between ADAMTS10 expression and the clinicopathological features of GC. Moreover, we aimed to elucidate the relationship between ADAMTS10 and TXNIP and the mechanisms of ADAMTS10 in GC.
Materials and methods
Patients and cancer tissue samples
A total of 333 primary GC tissue samples were collected at the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China, from December 2007 to March 2014. Three M0 patients and one M1 patient received R1 resection were excluded from this database, therefore, 329 patients received R0 resection were included and followed up until death or March 31, 2019. The interval between the date of surgery and the date of death or last follow-up was considered overall survival (OS). The American Joint Committee on Cancer Staging System (7th edition) was used for GC staging. Informed consent was obtained from all patients. Tissue specimens were constructed using immunohistochemical tissue microarrays (TMAs).
Immunohistochemistry
The Biotin-Streptavidin–Horseradish Peroxidase (HRP) Assay System (ZSGB Bio, Beijing, China) for immunohistochemistry (IHC) staining was used as described previously [27]. The TMAs were incubated with primary rabbit antibodies against ADAMTS10 (ab230902; 1:100; Abcam, Cambridge, UK). ADAMTS10 expression scores were assigned independently by two pathologists. X-tile software version 3.6.1 (Rimm Laboratories, New Haven, CT, USA) was then used to select the best cutoff score (6.1) [28]. Immunohistochemical analysis was performed on 34 pairs of primary GC and adjacent normal mucosal tissues to assess the differential expression of ADAMTS10. A TMA containing 329 primary cancer tissue specimens was used to assess the relationship between ADAMTS10 expression and the clinicopathological features and prognosis of GC. Based on the ADAMTS10 scores, the 329 patients were divided into two groups: high ADAMTS10 (score ≥ 6.1) and low ADAMTS10 (score < 6.1).
Public online database and related analyses
The public database TIMER2.0 (http://timer.cistrome.org; accessed September 25, 2021) was used to estimate the correlation between ADAMTS10 and macrophages in stomach adenocarcinoma.
Cell lines and cultures
Seven human GC cell lines (MKN1, MKN45, MGC803, BGC803, HGC27, SGC7901, and AGS), one human normal gastric mucosal cell line (GES1) and one human monocyte THP-1 cell line were obtained from the Type Culture Collection Cell Bank of the Chinese Academy of Sciences Committee (Shanghai, China). AGS was cultured in DMEM/F12, GES1 and HGC27 were cultured in DMEM, and the other cell lines were cultured in RPMI 1640. All medium was supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
THP-1 monocytes were differentiated into macrophages by incubation with 150 nM phorbol 12-myristate 13-acetate (Sigma, St. Louis, MO, USA) for 24 h followed by incubation in RPMI 1640 for 24 h, and then were cultured in condition medium(CM) for further experiments.
Conditioned medium and overexpression lysate protein
The ADAMTS10 overexpression cell lines AGS and HGC27 and the ADAMTS10 knockdown cell lines MKN1 and SGC7901 were cultured in a serum-free medium for 48 h. The supernatant was collected as Conditioned medium (CM), and cell debris was removed by centrifugation at 14,000g for 10 min. Over-Expression Lysate Protein (LS-G92301-20, LSBio, Seattle, WA, USA) was diluted according to the manufacturer’s protocol and added into CM depending on the design of experiments.
Plasmid construction and transfection for gene silencing and expression
The full-length complementary DNA (cDNA) encoding ADAMTS10, TXNIP, and c-MYC was amplified from the complete open reading frame of ADAMTS10 (NM_030957.4), TXNIP (NM_006472), and c-MYC (NM_001354870.1). ADAMTS10 cDNA was then cloned into pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro vector with a 3 × FLAG tag. Other plasmids, such as pCDNA3.1ADAMTS10-3 × FLAG, pCDNA3.1TXNIP-3 × FLAG, and pCDNA3.1c-MYC-3 × FLAG were constructed using the same method. For ADAMTS10 knockdown, an ADAMTS10 single guide RNA (sgRNA) lentiviral vector and a control sgRNA vector were purchased from Genechem (Shanghai, China). The target sequences of ADAMTS10 sgRNA were 1S: 5′-caccGGCGTCGCACGAGTGCACGG-3′; 1AS: 5′-aaacCCGTGCACTCGTGCGACGCC-3′; 2S: 5′-caccGAAGAGGCGGGACTCGGCTG-3′, 5′-aaacCAGCCGAGTCCCGCCTCTTC-3′.
For pCDNA3.1 plasmids, transient transfection was performed using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). Small interfering RNA (siRNA) for ADAMTS10, TXNIP and c-MYC (RiboBio, Guangzhou, China) was transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA). The targeting sequences of siRNA were SiADAMTS10-1: 5′-CCGGGAAGCACCGTTAACATCCTCTCGAGAGGATGTTAACGGTGCTTCTTTTTT-3′ (forward); SiADAMTS10-2: 5′-CCGGGGAGTCAAATCGCGTCAGTCTCGAGACTGACGCGATTTGACTCCTTTTTT-3′ (forward); SiTXNIP-1: 5′-GAAACAAAUAUGAGUACAA-3′ (forward); SiTXNIP-2: 5′-GAGAAUACAUGUUCCCGAA-3′ (forward); Sic-MYC: 5′-CCTGAGACAGATCAGCAACAA-3′ (forward) [29].
Western blot analysis
Cellular protein was extracted with T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Massachusetts, MA, USA). Equivalent amounts of protein extracts were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore, Massachusetts, MA, USA). The membranes were then blocked with 5% skimmed milk at room temperature for 1 h and incubated with primary antibodies (Supplementary Table 1) at 4 °C overnight. This was followed by incubation with the corresponding HRP-conjugated secondary antibody (goat anti-mouse IgG SA00001-1, 1: 10,000, Proteintech, Wuhan, China; mouse anti-rabbit IgG light chain specific SA00001-7L, 1: 10,000, Proteintech, Wuhan, China) at room temperature for 1 h. Bands were then visualized using ChemiDoc Touch imaging system (Bio-Rad, California, CA, USA). The bands were quantified using ImageJ software version 1.52v (National Institutes of Health, Bethesda, MD, USA) and standardized using tubulin.
RNA extraction and qRT-PCR
Total RNA was extracted from tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and from cells using an RNA Rapid Purification Kit (ES-RN001; Yishan Biotechnology, Shanghai, China). Reverse transcription PCR and qRT-PCR were performed as described previously [30]. To assess ADAMTS10, 29 pairs of freshly frozen primary cancer tissue samples and paracancerous normal tissue samples were used to assess the levels of messenger RNA (mRNA).
The qRT-PCR primer sequences were: ADAMTS10, 5′-AGAAACCAAGGCCCAGAGAG-3′ (forward) and 5′-ATCACCGGGCTCCTTTTGG-3′ (reverse); GAPDH, 5′-GACAGTCAGCCGCATCTTCTT-3′ (forward) and 5′-AATCCGTTGACTCCGACCTTC-3′ (reverse); TXNIP, 5′-GGTCTTAACGACCCTGAAAAGG-3′ (forward) and 5′-ACACGAGTAACTTCACACACCT-3′ (reverse); CD163, 5′-TTTGTCAACTTGAGTCCCTTCAC-3′ (forward) and 5′-TCCCGCTACACTTGTTTTCAC-3′ (reverse); Arg1, 5′-TGGACAGACTAGGAATTGGCA-3′ (forward) and 5′-CCAGTCCGTCAACATCAAAACT-3′ (reverse).
Migration and invasion assays
Transwell chambers (353,097; Falcon, New York, NY, USA) were used with or without Matrigel (356,234; Corning, New York, NY, USA) to assess GC cell migration and invasion. Briefly, 4 × 104 cells were transferred to the upper chamber, containing 100 µL of serum-free medium, while the lower chamber was injected with 700 µL of 10% serum medium to produce a chemoattractant. After sufficient incubation, the cells on the beneath surface of the upper membrane were fixed using 4% paraformaldehyde and stained with crystal violet. Five random fields on the upper membrane surface were photographed using a microscope (Olympus, Tokyo, Japan), and the number of migrating and invading cells was calculated using ImageJ (National Institutes of Health, Bethesda, MD, USA).
Wound healing assay
After inoculating 1 × 106 cells in a 12-well plate, wounds were made using the sterile tips of 200-µL pipettes. The cells were then incubated with a 1% serum culture medium. Images of the wound area were acquired every 2 h during incubation using an Incucyte ZOOM System (Essen BioSciences, Michigan, MI, USA). The initial wound area was defined 100%, and the percentage of wound closure at the end of the culture process was calculated using ImageJ.
Cell apoptosis and cycle assays
Cells were seeded into a 6-well plate and cultured overnight to adhere. Upon growing to cover 80–90% of the plate, they were collected and stained using an Annexin V-APC/7-AAD Apoptosis Kit (MultiSciences Biotech, Hangzhou, China) or a cell cycle staining kit (MultiSciences Biotech, Hangzhou, China), and then examined using a flow cytometer (CytoFLEX S, Beckman Coulter, Baker, Montana, USA).
Management and measurement of ROS generation in cells
We used N-acetyl-L-cysteine (NAC) to decreased ROS of culture cells and hydrogen peroxide (H2O2) to increased ROS of culture cells.
Cellular ROS production was measured by flow cytometry using a ROS-sensitive dye. Briefly, 5 × 105 cells were inoculated in 6-well culture dishes in a normal growth medium with or without N-acetyl-L-cysteine (NAC) or hydrogen peroxide (H2O2). The cells were then stained with 10 μM DHE (UElandy, Suzhou, China) at 37 °C for 30 min, avoiding light. The ROS levels were measured using a flow cytometer (CytoFLEX S, Beckman Coulter, Baker, Montana, USA).
RNA sequencing array and bioinformatics analysis
Whole-transcript deep sequencing (RNAseq) was performed on a BGISEQ-500 platform (BGI, Shenzhen, China). The analysis was designed using two groups of paired AGS vector/ADAMTS10 and SGC7901–SgNC/SgADAMTS10 cells. Heat map analysis of altered genes was performed using the Sangerbox heat map tool (http://vip.sangerbox.com/home.html; accessed September 3, 2021). Gene set enrichment analysis (GSEA) was performed using the GSEA preprocessing tool, R version 4.0.3 (R Foundation, New York, NY, USA), and RStudio version 1.4.1103 (RStudio, Boston, MA, USA).
Tumor xenotransplantation model
AGS cells (1 × 106) transfected with ADAMTS10 and its control vectors were stably injected subcutaneously into the left and right sides of female BALB/c nude mice, respectively, (n = 6; 3–4 weeks old; purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd, Beijing, China). The tumor volumes were measured on the 5th, 10th, 15th, 20th, and 27th days after injection. On day 27, the mice were euthanized by cervical dislocation. The tumor volumes (in cubic mm) and masses (in g) were measured. The tumor volume was calculated according to the following formula: volume = length × width2 × 0.5.
IHC and TUNEL analyses were performed on the collected subcutaneous tumors. All animal experiments were approved by the Ethics Committee of the Sixth Affiliated Hospital of Sun Yet-sen University. Every effort was made to minimize the suffering of the mice.
TUNEL assay
Mouse tumor tissue samples were embedded in paraffin, sectioned, and subjected to TUNEL analysis using an In Situ Cell Death Detection Kit (Roche, Basel, Switzerland) according to the manufacturer’s protocol. The samples were incubated with fluorescent secondary antibodies at room temperature for 2 h. Cell nuclei were stained with DAPI. Digital images of the cells were obtained using a TCS SP8 microscope (Leica, Wetzlar, Germany).
Statistical analysis
IBM SPSS Statistics Version 21.0 (IBM, New York, NY, USA) and GraphPad Prism (GraphPad Software, San Diego, CA, USA) were used for statistical analysis. Continuous variables were assessed using Student’s t test and reported as means ± standard deviations. Categorical variables were analyzed using the chi-squared test or the Wilcoxon signed-rank test as appropriate. Overall survival was calculated using the Kaplan–Meier method and compared between the high and low ADAMTS10 score groups using the log-rank test. Cox proportional hazards regression was used to assess the potential prognostic factors of survival in multivariate analyses during backward elimination. P values less than 0.05 were considered statistically significant.
Results
ADAMTS10 is downregulated in human GC
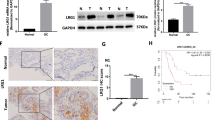
ADAMTS10 mRNA levels were significantly lower in cancerous tissue than in normal tissue. Similar results were obtained by qRT-PCR on 29 pairs of cancerous and normal tissues (Fig. 1a) and IHC on 34 pairs of cancerous and normal tissues (Fig. 1b). Representative IHC images are shown in Fig. 1c.
ADAMTS10 was decreased in human gastric cancer (GC). a Quantitative real-time polymerase chain reaction (qRT-PCR) on 29 paired samples showed that ADAMTS10 mRNA expression levels were lower in tumor tissue than in adjacent normal tissue. b Immunohistochemistry (IHC) analysis of 34 paired samples showed that ADAMTS10 expression was lower in tumor tissue than in adjacent normal tissue. c Representative IHC images of tumor and adjacent normal tissues. d High ADAMTS10 expression predicted better overall survival than low expression in GC patients (the best cutoff score of 6.1 was obtained using X-tile). e, f ADAMTS10 protein expression levels measured in seven GC cell lines (MKN1, MGC803, BGC823, HGC27, SGC7901, and AGS) and one normal gastric epithelial cell line (GES1) by western blotting and qRT-PCR. g, h HGC27 and AGS cells with low endogenous ADAMTS10 expression were chosen to construct stably overexpressed ADAMTS10 cells transfected with a lentiviral vector, and MKN1 and SGC7901 cell lines with high endogenous ADAMTS10 expression were stably transfected with sgRNA using a lentiviral vector. ADAMTS10 expression in stably transfected cells was confirmed by western blotting and qRT-PCR. The data are presented as means ± standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
High expression of ADAMTS10 is associated with better OS in GC
We evaluated the correlations between ADAMTS10 expression and clinicopathological features in 329 GC samples. ADAMTS10 protein expression did not significantly correlate with age, gender and histological type. ADAMTS10 protein expression significantly correlate with differentiation, T stage, N stage, M stage, TNM stage, perineural invasion and vessel invasion (Table 1). Survival analysis of 329 patients showed significantly better OS among patients with high ADAMTS10 expression than among those with low expression (Fig. 1d). Multivariate analysis identified four prognostic indicators of survival: invasion depth, lymph node metastasis, distant metastasis and ADAMTS10 expression. ADAMTS10 was an independent prognostic factor in multivariate analysis (HR = 0.338, p < 0.001, Table 2).
ADAMTS10 inhibits GC cell proliferation, migration, and invasion, promotes apoptosis, and alters the cell cycle
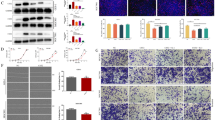
We identified the expression of ADAMTS10 in GC cell lines and found that ADAMTS10 was low endogenous expressed in AGS, HGC27 and MGC803, and was high endogenous expression expressed in MKN1, SGC7901 and BGC823. (Fig. 1e, f). We transfected AGS and HGC27 with lentiviral vectors to induce stable ADAMTS10 overexpression. We transfected MKN1 and SGC7901 with lentiviral vectors to induce stable ADAMTS10 knockdown (Fig. 1g, h).
Colony formation assays showed that ADAMTS10 ectopic expression inhibited cell growth (Fig. 2a). Proliferation assays showed that ADAMTS10 inhibited cell proliferation (Fig. 2c1; Supplementary Fig. 1a1). Cells with ADAMTS10 overexpression had increased G0/G1 phase cells and decreased G2/M and S phase cells (Fig. 2d and Supplementary Fig. 1b). Flow cytometry showed that ADAMTS10 promoted apoptosis (Fig. 2e). Migration assays showed that ADAMTS10 ectopic expression inhibited cell migration (Fig. 2f). Similarly, invasion analysis showed that ADAMTS10 ectopic expression inhibited cell invasion (Fig. 2g). Wound healing assays showed that ADAMTS10 overexpression inhibited wound healing (Fig. 2h and Supplementary Fig. 1c). In contrast, ADAMTS10 knockdown promoted cell proliferation, migration, invasion, and wound healing, inhibited apoptosis, decreased G0/G1 phase cells, and increased G2/M and S phase cells (Fig. 2b, c2, Supplementary Fig. 1a2, d–h). The results of the in vivo experiment were similar to the in vitro results. Vector tumors grew faster and larger than ADAMTS10 tumors (Fig. 2i1–i3 and Supplementary Fig. 1i). Ki67 analysis showed that ADAMTS10 inhibited tumor cell proliferation, and TUNEL assays showed that ADAMTS10 promoted tumor cell apoptosis (Fig. 2i4).
ADAMTS10 inhibited GC cell proliferation, migration, and invasion, promoted apoptosis, and altered the cell cycle in vitro and in vivo. a–c Colony and proliferation assays showed that ADAMTS10 overexpression suppressed GC cell proliferation, whereas ADAMTS10 knockdown promoted GC cell proliferation. d Cell cycle assays showed that cells with ADAMTS10 overexpression had increased G0/G1 phase cells and decreased G2/M and S phase cells. e Apoptosis assays showed that ADAMTS10 overexpression increased GC cell apoptosis. f, g ADAMTS10 overexpression suppressed migration and invasion. h Wound healing assays showed that ADAMTS10 overexpression significantly inhibited wound healing in AGS and HGC27. i In vivo experiments showed that ADAMTS10 overexpression inhibited GC cell proliferation and promoted apoptosis. The data are presented as means ± standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
TXNIP is downstream of ADAMTS10 and inhibits tumor proliferation and invasion
Heat map analysis based on RNAseq revealed distinct genes after ADAMTS10 overexpression and knockdown (Fig. 3a; for the genes involved, see Supplementary Table 2). HALLMARK_INTERFERON_GAMMA_RESPONSE and other pathways were enriched in GSEA (Fig. 3b). Among the positively correlated genes, TXNIP stood out for its highly correlation with ADAMTS10 and greatly change according to ADMATS10. Western blotting and qRT-PCR showed that TXNIP expression was influenced by ADAMTS10 expression (Fig. 3c). In contrast, western blotting showed that TXNIP expression did not affect ADAMTS10 expression (Fig. 3d). Western blotting and IHC showed that TXNIP was positive correlated with ADAMTS10 in GC cell lines and GC specimens (Supplementary Fig. 2a, b). We also investigated the role of TXNIP using in vitro functional assays. TXNIP overexpression inhibited cell proliferation (Fig. 3e) and invasion (Fig. 3f), which were promoted by ADAMTS10 downregulation. Conversely, TXNIP silencing reversed the suppression effect on cell proliferation (Fig. 3g) and invasion (Fig. 3h) by ADAMTS10 overexpression.
TXNIP is a downstream factor of ADAMTS10 and acts as a tumor suppressor, inhibiting cell proliferation and invasion. a Heat map analysis showed altered genes in AGS-Vector, AGS-ADAMTS10, SGC7901-SgNC, and SGC7901-SgADAMTS10-1 cells. b Gene set enrichment analysis showed the enrichment of ADAMTS10-associated genes in HALLMARK_INTERFERON_GAMMA_RESPONSE and other pathways. c Quantitative RT-PCR and western blotting showed that TXNIP mRNA and protein levels were affected by ADAMTS10. d TXNIP ectopic expression and knockdown did not affect the expression of ADAMTS10. e, f TXNIP knockdown reversed the inhibition of cell proliferation and invasion by ADADAMTS10 in AGS and HGC27 cells. g, h TXNIP ectopic expression reversed the promotion of cell proliferation and invasion by ADAMTS10 knockdown in MKN1 and SGC7091 cells. The data are presented as means ± standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
ADAMTS10 inhibits GC cell proliferation and invasion via ROS regulation through the JAK/STAT/c-MYC/TXNIP axis
We further explored the potential ADAMTS10–TXNIP mechanism. ADAMTS10 overexpression downregulated JAK1, JAK2, STAT1, STAT3, and c-MYC, whereas ADAMTS10 knockdown significantly upregulated them (Fig. 4a). Moreover, TXNIP was upregulated and downregulated when c-MYC was silenced and overexpressed, respectively (Fig. 4b). The changes in TXNIP expression were followed by changes in ROS levels. In ADAMTS10 overexpression cells, TXNIP was elevated, and ROS levels were also increased (Fig. 4c). Conversely, TXNIP silencing resulted in decreased ROS levels (Fig. 4d). In ADAMTS10 knockdown cells, TXNIP was reduced, and ROS levels were also decreased. In contrast, TXNIP overexpression resulted in increased ROS levels (Supplementary Fig. 2a, b). Next, we increased and decreased the ROS levels (Fig. 4e, Supplementary Fig. 2c). Elevated ROS levels prevented GC cell proliferation (Fig. 4f) and invasion (Fig. 4g). Conversely, reduced ROS levels promoted GC cell proliferation and invasion (Supplementary Fig. 2d, e).
ADAMTS10 upregulated TXNIP by inhibiting the JAK/STAT/c-MYC pathway. ADATMTS10 inhibited GC cell proliferation and invasion via the JAK/STAT/c-MYC/TXNIP/ROS axis. a Western blotting showed the JAK/STAT/c-MYC pathway were affected by ADAMTS10. b Western blotting showed that TXNIP protein levels were affected by c-MYC. c Flow cytometry showed that ROS levels were affected by ADAMTS10 in AGS and HGC27 cells. d Flow cytometry showed that ROS levels were affected by TXNIP in AGS, HGC27, MKN1, and SGC7901 cells. e–g Colony and invasion assays showed that decreased ROS levels reversed the inhibition of cell proliferation and invasion by ADAMTS10 in AGS and HGC27 cells. The data are presented as means ± standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
ADAMTS10 secreted by GC cells alters ROS in macrophages and inhibits macrophage M2 polarization
We explored the relationship between ADAMTS10 and macrophages using TIMER2.0 and found that ADAMTS10 was positively related to macrophages (Fig. 5a). Next, we cultured THP1 with CM collected from the ADAMTS10 overexpression cell lines AGS and HGC27. We found that Flag–ADAMTS10 appeared in THP1, suggesting that ADAMTS10 secreted by GC cells was absorbed by THP1 (Fig. 5b), resulting in increased THP1 TXNIP (Fig. 5b, c) and ROS levels (Fig. 5d). In contrast, ROS levels decreased when we cultured THP1 with a supernatant collected from the ADAMTS10 knockdown cell lines MKN1 and SGC7901 (Fig. 5e). Furthermore, we increased and decreased the expression of ADAMTS10 in THP1 and found that ADAMTS10 inhibited the JAK/STAT/c-MYC pathway and increased the expression of TNXIP (Fig. 5f), thereby raising the levels of ROS (Fig. 5g). When THP1 was cultured in CM with exogenous ADAMTS10 protein, JAK/STAT/c-MYC pathway in it was also inhibited and TXNIP was also increased (Fig. 5h, i). Moreover, we found that ADAMTS10 from GC cells downregulated CD163 and Arg1 in THP1, suggesting that it can inhibit macrophage M2 polarization (Fig. 5j, Supplementary Fig. 3a). The same phenomenon was observed when THP1 culture in CM with exogenous ADAMTS10 protein (Fig. 5k, Supplementary Fig. 3b).
ADAMTS10 secreted by GC cells altered ROS through the JAK/STAT/c-MYC/TXNIP axis in macrophages and inhibited macrophage M2 polarization. a The positive correlation between ADAMTS10 and macrophages was predicted using TIMER2.0. The red indicates a statistically significant positive association, the blue indicates a statistically significant negative association, and the gray denotes a non-significant result. The EPIC, TIMER and MCP-COUNTER algorithms predicted that the expression of ADAMTS10 was positive associated with macrophage infiltration. The CIBERSORT-ABS algorithm predicted that the expression of ADAMTS10 was positive associated with M0, M1 and M2 macrophage infiltration. The CIBERSORT and QUANTISEQ algorithms predicted that overexpression of ADAMTS10 was positive associated with M2 macrophage infiltration. b Western blotting showed that ADAMTS10 secreted by GC cells was absorbed by THP1 and affected the expression of TXNIP in THP1. c Quantitative RT-PCR analysis showed that ADAMTS10 affected the expression of TXNIP in THP1. d, e Flow cytometry showed that ADAMTS10 secreted by GC cells affected the ROS levels in THP1. f Western blotting showed that ectopic expression and knockdown of ADAMTS10 in THP1 altered the JAK/STAT/c-MYC/TXNIP axis. g Flow cytometry showed that ADAMTS10 ectopic expression and knockdown altered the ROS levels in THP1. h Western blotting showed that exogenous ADAMTS10 protein inhibited JAK/STAT/c-MYC pathway and increased TXNIP in THP1. i Quantitative RT-PCR showed that exogenous ADAMTS10 protein increased TXNIP in THP1. j Quantitative RT-PCR analysis showed that ADAMTS10 secreted by GC cells inhibited macrophage M2 polarization. k Quantitative RT-PCR analysis showed that exogenous ADAMTS10 protein inhibited macrophage M2 polarization. The data are presented as means ± standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Discussion
The ADAMTS family has been shown to be related to several cancers. ADAMTS genes can act as either cancer promoters or inhibitors. For instance, ADAMTS1 is overexpressed in pancreatic cancer and fibrosarcoma and is involved in promoting tumor growth and invasion [31]. ADAMTS4 can increase the invasion of glioblastoma cancer cells and head and neck squamous cell carcinoma [32]. ADAMTS5 can act as a tumor suppressor by inhibiting angiogenesis in human GC [33]. ADAMTS9 has also been found to act as a tumor suppressor by inhibiting angiogenesis [34]. High ADAMTS15 expression is associated with better relapse-free survival and may prevent colorectal and breast cancer metastasis [35]. ADAMTS19 suppresses GC via the NF-κB pathway [30]. Previous studies on ADAMTS10 have focused on its relationship with genetic diseases [14, 15, 36, 37], but none have investigated its relationship with cancers. This is the first study to explore its role in GC.
Using qRT-PCR and IHC, we found that ADAMTS10 expression was downregulated in GC tissue compared to adjacent normal tissue. ADAMTS10 protein was associated with with differentiation, T stage, N stage, M stage, TNM stage, perineural invasion and vessel invasion in GC. Survival analysis showed that patients with low ADAMTS10 expression had worse OS than those with high expression. Overall, the results suggest that ADMTS10 may be a GC prognostic marker. Cell function analysis showed that high ADAMTS10 inhibited GC cell migration, invasion, and proliferation, promoted cell apoptosis, increased G0/G1 phase cells, and decreased G2/M and S phase cells. We also demonstrated the tumor growth–inhibiting effect of ADAMTS10 in vivo. Previous studies have shown that JAK/STAT/c-MYC is a cancer-promoting pathway and is associated with tumor proliferation and invasion [38,39,40]. However, the relationship between ADAMTS10 and the JAK/STAT/c-MYC pathway has not been previously investigated. Although some studies have used bioinformatics to determine the association between TXNIP and the JAK/STAT pathway, no studies have directly examined whether TXNIP is upstream or downstream of the JAK/STAT pathway [40]. Several studies have reported that c-MYC is an upstream molecule of TXNIP and can regulate TXNIP expression [41, 42]. In this study, we found that ADAMTS10 inhibited the JAK/STAT/c-MYC pathway, thereby upregulating TXNIP.
Although evidence suggests that TXNIP is associated with ROS, it is not so clear which one is upstream of which. In the KEGG pathway map (https://www.kegg.jp/pathway/map04621+K20910; accessed September 25, 2021), TXNIP is considered downstream of ROS [43], while a previous study found that TXNIP acted as an upstream factor of ROS and regulated its levels [44]. In this study, we increased and decreased the expression of ADAMTS10 and observed alterations in TXNIP and ROS. Through rescue experiments, we confirmed that TXNIP is an upstream molecule of ROS and that ADAMTS10 regulates downstream ROS through TXNIP and inhibits tumor proliferation and invasion by upregulating ROS.
We investigated the relationship between ADAMTS10 and macrophages using TIMER2.0. Previous studies have shown that ADAMTS10 degrades FBN extracellularly [11, 12]. However, the role of ADAMTS10 in immune cells has not been previously documented. Our experiments demonstrate for the first time the effect of ADAMTS10 on macrophages, showing that ADAMTS10 secreted by GC cells can enter THP1 and regulate downstream TXNIP through the JAK/STAT/c-MYC pathway, affecting the levels of ROS. Our findings also show that ADAMTS10 can inhibit macrophage M2 polarization.
To our knowledge, our study not only reveals for the first time the role of ADAMTS10 and TXNIP in GC but also elucidates for the first time the regulatory mechanism of ADAMTS10–TXNIP. Next, we plan to conduct a direct mechanistic study on the role of ADAMTS10 in regulating the JAK/STAT/c-MYC pathway and modulating TXNIP to further explore its role in GC cell migration and invasion. Moreover, we plan to further investigate the specific mechanism of ROS elevation and M2 polarization inhibition in THP driven by ADAMTS10.
Conclusions
In this study, we found that ADAMTS10 expression was downregulated in GC and correlated with the prognosis of GC. Patients with elevated ADAMTS10 had better OS. In vitro, ADAMTS10 functionally altered the cell cycle, promoted apoptosis, and inhibited proliferation, migration, and invasion. Mechanistically, ADAMTS10 regulates TXNIP and ROS via the JAK/STAT/c-MYC pathway, thereby inhibiting cell proliferation and invasion. ADAMTS10 secreted by GC cells could enter THP1, affecting its ROS level and inhibiting the M2 polarization of macrophages. The clinical and biological significance of the ADAMTS10/JAK/STAT/c-MYC/TXNIP/ROS axis suggests that ADAMTS10 could be used as a prognostic biomarker of GC.
References
Smyth EC, Nilsson M, Grabsch HI, van Grieken NCT, Lordick F. Gastric cancer. Lancet. 2020;396:635–48.
Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113.
Yang CY, Chanalaris A, Troeberg L. ADAMTS and ADAM metalloproteinases in osteoarthritis – looking beyond the ‘usual suspects.’ Osteoarthritis Cartilage. 2017;25:1000–9.
Cal S, Lopez-Otin C. ADAMTS proteases and cancer. Matrix Biol. 2015;44–46:77–85.
Le Goff C, Cormier-Daire V. The ADAMTS(L) family and human genetic disorders. Hum Mol Genet. 2011;20:R163–7.
Sun Y, Huang J, Yang Z. The roles of ADAMTS in angiogenesis and cancer. Tumour Biol. 2015;36:4039–51.
Wagstaff L, Kelwick R, Decock J, Edwards DR. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front Biosci-Landmark. 2011;16:1861–72.
Hubmacher D, Apte SS. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 2015;47:34–43.
Mead TJ, Apte SS. ADAMTS proteins in human disorders. Matrix Biol. 2018;71–72:225–39.
Cain SA, Mularczyk EJ, Singh M, Massam-Wu T, Kielty CM. ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci Rep. 2016;6:35956.
Wang LW, Kutz WE, Mead TJ, Beene LC, Singh S, Jenkins MW, et al. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 2019;77:117–28.
Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, et al. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J Biol Chem. 2011;286:17156–67.
Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Mégarbané A, Alswaid A, et al. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am J Hum Genet. 2004;75:801–6.
Kutz WE, Wang LW, Dagoneau N, Odrcic K, Cormier-Daire V, Traboulsi EI, et al. Functional analysis of an ADAMTS10 signal peptide mutation in Weill-Marchesani syndrome demonstrates a long-range effect on secretion of the full-length enzyme. Hum Mutat. 2008;29:1425–34.
Morales J, Al-Sharif L, Khalil DS, Shinwari JMA, Bavi P, Al-Mahrouqi RA, et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85:558–68.
Xiaoyi H, Jing L, Maorong F, Xia Z, Wei W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transd Targeted Ther. 2021;6:402.
Ludwig DL, Kotanides H, Le T, Chavkin D, Bohlen P, Witte L. Cloning, genetic characterization, and chromosomal mapping of the mouse VDUP1 gene. Gene. 2001;269:103–12.
Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, et al. Identification of thioredoxin-binding protein-2/vitamin D3 up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–50.
Xie M, Xie R, Xie S, Wu Y, Wang W, Li X, et al. Thioredoxin interacting protein (TXNIP) acts as a tumor suppressor in human prostate cancer. Cell Biol Int. 2020;44:2094–106.
Chung JW, Jeon JH, Yoon SR, Choi I. Vitamin D3 upregulated protein 1 (VDUP1) is a regulator for redox signaling and stress-mediated diseases. J Dermatol. 2006;33:662–9.
Hwang J, Suh HW, Jeon YH, Hwang E, Nguyen LT, Yeom J, et al. The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein. Nat Commun. 2014;5:2958.
Kim SY, Suh H-W, Chung JW, Yoon S-R, Choi I. Diverse functions of VDUP1 in cell proliferation, differentiation, and diseases. Cell Mol Immunol. 2007;4(5):345–51.
Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49:1167–75.
Jeon J-H, Lee K-N, Hwang CY, Kwon K-S, You K-H, Choi I, et al. Tumor suppressor VDUP1 increases p27kip1 stability by inhibiting JAB1. Cancer Res. 2005;65(11):4485–9.
Li J, Yue Z, Xiong W, Sun P, You K, Wang J. TXNIP overexpression suppresses proliferation and induces apoptosis in SMMC7221 cells through ROS generation and MAPK pathway activation. Oncol Rep. 2017;37:3369–76.
Su C, Shi A, Cao G, Tao T, Chen R, Hu Z, et al. Fenofibrate suppressed proliferation and migration of human neuroblastoma cells via oxidative stress dependent of TXNIP upregulation. Biochem Biophys Res Commun. 2015;460:983–8.
Huang J, Bai Y, Huo L, Xiao J, Fan X, Yang Z, et al. Upregulation of a disintegrin and metalloprotease 8 is associated with progression and prognosis of patients with gastric cancer. Transl Res. 2015;166(6):602–13.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–9.
Ji S, Qin Y, Liang C, Huang R, Shi S, Liu J, et al. FBW7 (F-box and WD repeat domain-containing 7) negatively regulates glucose metabolism by targeting the c-Myc/TXNIP (thioredoxin-binding protein) axis in pancreatic cancer. Clin Cancer Res. 2016;22:3950–60.
Jiang Y, Yu X, Zhao Y, Huang J, Li T, Chen H, et al. ADAMTS19 suppresses cell migration and invasion by targeting S100A16 via the NF-κB pathway in human gastric cancer. Biomolecules. 2021;11(4):561.
Casal C, Torres-Collado AX, Plaza-Calonge Mdel C, Martino-Echarri E, Ramon YCS, Rojo F, et al. ADAMTS1 contributes to the acquisition of an endothelial-like phenotype in plastic tumor cells. Cancer Res. 2010;70:4676–86.
Demircan K, Gunduz E, Gunduz M, Beder LB, Hirohata S, Nagatsuka H, et al. Increased mRNA expression of ADAMTS metalloproteinases in metastatic foci of head and neck cancer. Head Neck. 2009;31:793–801.
Huang J, Sun Y, Chen H, Liao Y, Li S, Chen C, et al. ADAMTS5 acts as a tumor suppressor by inhibiting migration, invasion and angiogenesis in human gastric cancer. Gastric Cancer. 2019;22:287–301.
Clark ME, Kelner GS, Turbeville LA, Boyer A, Arden KC, Maki RA. ADAMTS9, a novel member of the ADAM-TS/metallospondin gene family. Genomics. 2000;67:343–50.
Porter S, Span PN, Sweep FC, Tjan-Heijnen VC, Pennington CJ, Pedersen TX, et al. ADAMTS8 and ADAMTS15 expression predicts survival in human breast carcinoma. Int J Cancer. 2006;118:1241–7.
Pimienta AL, Wilcox WR, Reinstein E. More than meets the eye: the evolving phenotype of Weill–Marchesani syndrome—diagnostic confusion with geleophysic dysplasia. Am J Med Genet A. 2013;161A:3126–9.
Kochhar A, Kirmani S, Cetta F, Younge B, Hyland JC, Michels V. Similarity of geleophysic dysplasia and Weill–Marchesani syndrome. Am J Med Genet A. 2013;161A:3130–2.
Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106.
Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6:402.
Gao Y, Qi JC, Li X, Sun JP, Ji H, Li QH. Decreased expression of TXNIP predicts poor prognosis in patients with clear cell renal cell carcinoma. Oncol Lett. 2020;19:763–70.
Qu X, Sun J, Zhang Y, Li J, Hu J, Li K, et al. c-Myc-driven glycolysis via TXNIP suppression is dependent on glutaminase-MondoA axis in prostate cancer. Biochem Biophys Res Commun. 2018;504:415–21.
Shen L, O’Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL, et al. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci U S A. 2015;112:5425–30.
Zhang X, Zhao S, Yuan Q, Zhu L, Li F, Wang H, et al. TXNIP, a novel key factor to cause Schwann cell dysfunction in diabetic peripheral neuropathy, under the regulation of PI3K/Akt pathway inhibition-induced DNMT1 and DNMT3a overexpression. Cell Death Dis. 2021;12:642.
Chen D, Dang B-L, Huang J-Z, Chen M, Wu D, Xu M-L, et al. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget. 2015;6:32701–12.
Acknowledgements
This work was supported by grants from the National Key Clinical Discipline, the National Natural Science Foundation of China (Grant Nos 81772594, 81802322, and 81902949), the Science and Technology Program of Guangzhou (Grant No. 201803010095), and the Natural Science Foundation of Guangdong Province, China (Grant Nos 2020A1515011362 and 2022A1515010262).
Author information
Authors and Affiliations
Contributions
All authors contributed to concept elaboration and literature search strategy. JZ and ZY wrote the original draft of the article. Finally, all authors have provided critical review of the manuscript and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests in this study.
Ethical approval
Animal experiments were approved and performed in accordance with the institutional guidelines for animal care of animal ethics committee of Sun Yat-sen University. All institutional and national guidelines for the care and use of laboratory animals were followed. The study was approved by the Research Ethics Committee of Sun Yat-sen University and conducted in accordance with the principles of the Declaration of Helsinki.
Informed consent
Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, J., Li, T., Chen, H. et al. ADAMTS10 inhibits aggressiveness via JAK/STAT/c-MYC pathway and reprograms macrophage to create an anti-malignant microenvironment in gastric cancer. Gastric Cancer 25, 1002–1016 (2022). https://doi.org/10.1007/s10120-022-01319-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01319-4