Abstract
Background
L1 and SAT-α are repetitive DNA elements that undergo demethylation in association with cancerization. Unlike L1 hypomethaylation, nothing is known regarding the prognostic implication of SAT-α hypomethylation alongside L1 hypomethaylaton in gastric cancers.
Methods
Formalin-fixed paraffin-embedded samples from 492 cases of advanced gastric cancer were analyzed to determine their L1 and SAT-α methylation status using pyrosequencing methylation assay.
Results
L1 and SAT-α methylation statuses were correlated with clinicopathological parameters, including survival. L1 or SAT-α methylation levels were lower in gastric cancers with venous invasion or nodal metastasis than those without. L1 methylation was lower in gastric cancers with lymphatic emboli than in those with no lymphatic emboli, but was higher in gastric cancers with perineural invasion than in those with no perineural invasion. Multivariate survival analysis revealed that both tumoral L1 and SAT-α hypomethylations were found to correlate independently with OS (HR = 1.477; 95% CI 1.079–2.021 and HR = 1.394; 95% CI 1.011–1.922, respectively) and RFS (HR = 1.477; 95% CI 1.090–2.001 and HR = 1.516; 95% CI 1.106–2.078, respectively). Combined L1 and SAT-α hypomethylation turned out to correlate independently with OS (HR = 2.003; 95% CI 1.268–3.164) and RFS (HR = 2.226; 95% CI 1.411–3.510).
Conclusion
Not only tumoral L1 hypomethylation, but also tumoral SAT-α hypomethylation was found to be independent prognostic parameters in patients with advanced gastric cancer. SAT-α methylation status can be used to further divide gastric cancers with L1 hypomethylation into subsets of gastric cancers with better and worse prognosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytosines in CpG dinucleotides are the methylation targets of DNA methyltransferases. In normal cells, the CpG sites of CpG islands located in promoter and 5′ exonal sequences are usually protected from DNA methylation, but CpG sites located in other genomic sequences, particularly repetitive DNA elements, are usually methylated. Repetitive DNA elements are DNA sequences with high copy numbers and comprise approximately 50% of the human genome [1]. The two major classes of repetitive DNA elements are interspersed repeats and tandem repeats. Interspersed repeats include long interspersed nucleotide elements-1 (L1) and Alu, which are repeated half-million times and a million times in the human genome, respectively. L1 and Alu are heavily methylated in normal cells, and these methylations are closely associated with the repression of their retrotransposon activity [2, 3]. High-copy tandem repeats include alpha-satellite (α-SAT), β-SAT, and SAT1, II, and III. Although these repeats are located in centromeres and juxtacentromeric regions and their RNA transcription is repressed by constitutive heterochromatin formation, satellite RNAs are expressed in some conditions, including stress and demethylation [4, 5].
Cancer cells feature contrasting DNA methylation changes, namely, regional CpG island hypermethylation and diffuse genomic hypomethylation. The latter genomic hypomethylation involves mainly repetitive DNA elements, including both interspersed and tandem repeats [6]. Gastric cancers are known to develop through progression of the lesion from chronic gastritis to intestinal metaplasia to gastric adenoma. Although the methylation levels of repetitive DNA elements are lower in the cancer stage than in precancerous lesions or conditions, DNA methylation behaviors differ during multistep gastric carcinogenesis depending on the types of repetitive DNA elements [7, 8]. Tumoral L1 hypomethylation has been demonstrated to be associated with shortened survival time in patients with gastric cancers [9, 10], which might be related to the increased frequency of venous invasion or lymphatic emboli in association with tumoral L1 hypomethylation [10]. However, the hypomethylation of satellite sequences has not been investigated in the context of its relationships with the clinicopathological features of gastric cancer, including prognosis, although not only interspersed repeats, but also tandem repeats are demethylated in gastric cancers [7].
In the present study, we analyzed archival tissue samples of advanced gastric cancer for their methylation status in L1 and SAT-α using pyrosequencing methylation assay and correlated tumoral L1 or SAT-α hypomethylation with clinicopathological features, including survival. In a previous study using paired fresh-frozen and formalin-fixed, paraffin-embedded (FFPE) tissue samples, it was found that formalin fixation leads to an artificial increase in the measured value of L1 methylation level and that prolonged heating of the extracted DNA samples before bisulfite modification helped reduce the discrepancy in the measured value of L1 methylation levels between paired fresh-frozen and FFPE tissue samples [11]. Thus, in the present study, we adopted prolonged heat treatment of the DNA samples obtained from FFPE tissue samples for the purpose of lessening an artificial deviation in the measured value of the L1 or SAT-α methylation level. Because both SAT-α hypomethylation and L1 hypomethylation are two phenomena of diffuse genomic hypomethylation, we attempted to identify whether both SAT-α hypomethylation and L1 hypomethylation or one of these types of hypomethylation alone is a prognostic marker for advanced gastric cancers.
Materials and methods
Tissue samples and clinicopathological analysis
We collected formalin-fixed paraffin-embedded (FFPE) tumor material from a consecutive series of advanced gastric cancer (T2–T4) patients operated in Seoul National University between January 2007 and December 2008. Main inclusion criteria for the retrospective patient selection were age over 18, adenocarcinoma histology, T2 or greater stage, and availability of FFPE cancer tissues. Patients were excluded if they refused to participate in the molecular study, or had a history of neoadjuvant therapy for gastric cancer or other malignancy (except for thyroid papillary carcinoma) within 5 years. This study was approved by Seoul National University Hospital Institute Review of Board (IRB No. 1312-051-542) and was conducted in accordance with the Declaration of Helsinki. Clinical and histological data were retrieved from the electronic medical record, including tumor subsites within the stomach, Lauren’s classification, histological type, lymphatic emboli, perineural invasion, venous invasion, and tumor-node-metastasis (TNM) stage (American Joint Committee on Cancer, 7th edition). We checked electronic medical records carefully and excluded patients who had received palliative intent surgery and patients who had a history of other malignancy within 5 years. For multiple synchronous gastric cancers, data were derived from higher T stage tumor or larger tumors if the synchronous tumors were of the same T stage.
Pyrosequencing methylation analysis
Through microscopic examination of all available glass slides, up to 1 cm2 samples from tumor areas with the highest tumor purity and most prevalent histological type in the individual case were marked. The corresponding areas on unstained tissue slides were manually scraped and collected into the microtubes containing tissue lysis buffer solution (50 mM Tris, 1 mM EDTA, pH 8.0 and 1% Tween-20) and proteinase K (3 mg/ml). The microtubes were kept at 55 °C for 24 h and then incubated at 95 °C for 30 min. After spinning down the solutions, the supernatants were transferred into newly labeled microtubes. DNA samples were subjected to bisulfite modification using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA). The modified DNA samples were PCR-amplified with the same oligonucleotide primers, which were designed by the Dr. Yang group for pyrosequencing methylation assays of L1 and SAT-α [12]. For the measurement of L1 methylation levels, a pyrosequencing assay was performed as described previously. For SAT-α, the percentage of C/(C + T) at the first three consecutive CpG sites was averaged and defined as the methylation level. To assess assay precision, we evaluated the reproducibility of bisulfite modification efficiency and PCR-pyrosequencing and the variability of methylation measurement using the same FFPE DNA sample as described previously [13]. We performed pyrosequencing on 24 PCR products of L1 and 24 PCR products of SAT-α and measured the level of L1 methylation on 4 CpG sites and the level of SAT-α methylation on three CpG sites. Data for L1 and SAT-α methylation levels for the pool of tumor DNA sample are summarized in Supplementary Tables 1 and 2. The mean of bisulfite-to-bisulfite (between-bisulfite treatment) standard deviation (SD) was 1.617 and 2.596 for L1 and SAT-α, respectively. The mean of run-to-run (between-PCR pyrosequencing run) SD was 1.728 and 2.258 for L1 and SAT-α, respectively.
Statistical analysis
To determine whether the methylation levels of L1 and SAT-α were normally distributed, a normality test was performed with Shapiro–Wilk’s W test. Methylation levels of L1 and SAT-α were found to be not normally distributed. For the comparison of means, we used both parametric and non-parametric tests. Student t test and Mann–Whitney test were performed for comparisons of two groups, whereas ANOVA and Kruskal–Wallis test were performed for comparisons of three or more groups. Recurrence-free survival was calculated from the date of surgery to the date of recurrence or the date of death (whichever came first). Kaplan–Meier log rank test was performed to compare overall-survival and recurrence-free survival times across groups. Hazard ratio (HR) was calculated using the Cox proportional hazard model, and baseline characteristics were adjusted for covariates which were found to be significant in univariate survival analysis: age (younger and older), tumor subsite (involving and not involving cardia), T category, N category, M category, lymphatic emboli, venous invasion and perineural invasion. A backward stepwise elimination was carried out with P = 0.05 as a threshold, to select variables for the final model. Statistical analysis was performed with SPSS software for Windows, version 25.0 (SPSS, Chicago, IL, USA).
Results
Patient characteristics
A total of 492 patients with advanced gastric carcinoma were included. The male-to-female ratio was 2.06:1 (332 males and 160 females), and the median age was 61 years (range 23–86 years). Tumor subsite according to the epicenter of tumor was lower one-third in 250, mid-third in 147, and upper one-third in 95. Approximately 29% (n = 142) of the cases involved upper-one-third. There were 63 stage I, 162 stage II, 209 stage III, and 58 stage IV carcinomas.
L1 or SAT-α methylation levels and clinicopathological features
There was a positive correlation between L1 and SAT-α methylation levels (Pearson’s rho = 0.559, P < 0.001). Patient age was negatively correlated with L1 or SAT-α methylation level (Pearson’s rho = − 0.189, P < 0.001; Pearson’s rho = − 0.216, P < 0.001). L1 or SAT-α methylation tended to be lower in male gastric cancers than in female gastric cancers (Table 1). However, the relationship between age and L1 or SAT-α methylation level contrasted between male and female patients: female patients exhibited no difference in the methylation level of L1 or SAT-α between < 62 and ≥ 62 years of age, whereas in male patients, L1 or SAT-α methylation was significantly lower in tumors from patients ≥ 62 years of age than in tumors from patients < 62 years of age (Table 2). L1 or SAT-α methylation level was closely associated with Lauren histology with the highest and lowest methylation levels in gastric cancers of the diffuse type and intestinal type, respectively. L1 or SAT-α methylation level was lower in gastric cancers with venous invasion, lymphatic emboli, or nodal metastasis than in gastric cancers without the respective one. L1 methylation tended to be higher in gastric cancers with perineural invasion than in gastric cancers with no perineural invasion, but this result did not reach statistical significance. However, no association was found between L1 or SAT-α methylation level and T or M stage. Of the molecular subtypes defined by MSI and EBV statuses, L1 methylation level was the highest in EBV+ gastric cancers and the lowest in EBV−/MSI-gastric cancers. However, there was no significant difference in the methylation level of SAT-α between different molecular subtypes.
L1 or SAT-α methylation levels and patient survival
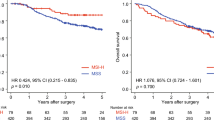
Gastric cancers were grouped into quintile subsets according to their L1 methylation level, and then, these quintile subsets were analyzed for their associations with recurrence-free survival (RFS) and overall survival (OS) using a Kaplan–Meier log rank test (Fig. 1a, b). Eighteen patients were excluded from survival analyses because of non-availability of survival data. Based on Kaplan–Meier survival analysis, five subsets were divided into two groups: a lower L1 methylation group (Q1, Q2 and Q3) and a higher L1 methylation group (Q4 and Q5) (Fig. 1c, d). The same approach was conducted for the survival analysis of SAT-α methylation level in patients with gastric cancer. Quintile subsets of gastric cancers according to tumoral SAT-α methylation level were analyzed for their RFS and OS by using Kaplan–Meier log rank test. RFS and OS were lower in subsets with lower SAT-α methylation levels (Q1, Q2, and Q3) than in subsets with higher SAT-α methylation levels (Q4 and Q5) (Fig. 2a–d). Then, multivariate analysis was performed to identify whether tumoral L1 or SAT-α hypomethylation was an independent prognostic value. Both L1 and SAT-α methylation statuses and other covariates of prognostic value on univariate survival analysis were incorporated into multivariate models. Tumoral L1 hypomethylation was found to correlate independently with OS (HR = 1.477; 95% CI 1.079–2.021) (Table 3) and RFS (HR = 1.477; 95% CI 1.090–2.001) (Table 3). Tumoral SAT-α hypomethylation was found to correlate independently with OS (HR = 1.394; 95% CI 1.011–1.922) (Table 3) and RFS (HR = 1.516; 95% CI 1.106–2.078) (Table 3).
Combinatory L1 and SAT-α hypomethylation as a prognostic factor
Although L1 and SAT-α are repetitive elements, L1 is interspersed over the chromosome, whereas SAT-α is localized to centromere. Because their methylation statuses represent different aspects of methylation statuses of the genome, combinatory methylation statuses of L1 and SAT-α are likely to better represent the methylation status of the whole genome than either the methylation status of L1 or SAT-α alone. Based on this plausibility, we attempted to identify whether combined L1 and SAT-α hypomethylation was a more powerful prognostic parameter than L1 hypomethylation or SAT-α hypomethylation alone. Of the four subsets generated by a combination of L1 and SAT-α methylation statuses, the subset with combined low L1 and SAT-α methylation statuses and the subset with combined high L1 and SAT-α methylation statuses showed the worst and best survival outcome, respectively, whereas the subset with low L1 and high SAT-α methylation statuses and the subset with high L1 and low SAT-α methylation statuses showed survival curves in-between (Fig. 3a, b). In multivariate analysis, combined L1 and SAT-α hypomethylation turned out to correlate independently with OS (HR = 2.004; 95% CI 1.269–3.166) and RFS (HR = 2.255; 95% CI 1.430–3.556) (Table 4). In both OS and RFS, the combinatory status of both L1 and SAT-α hypomethylation was found to have a higher hazard ratio than either L1 hypomethylation or SAT-α hypomethylation status alone.
Kaplan–Meier long rank tests of overall survival and recurrence-free survival in four subsets generated by a combination of L1 methylation status [low methylation status (Q1–Q3) vs. high methylation status (Q4 and Q5)] and SAT-α methylation status [low methylation status (Q1–Q3) vs. high methylation status (Q4 and Q5)]
Discussion
In the present study, we have analyzed archival tissue samples of advanced gastric cancers for their methylation statuses in L1 and SAT-α using pyrosequencing and correlated L1 and SAT-α methylation statuses with clinicopathological features, including prognosis. We found that both L1 hypomethylation and SAT-α hypomethylation were independent prognostic parameters heralding poor prognosis in both OS and RFS. Combinatory statuses of L1 and SAT-α methylation were found to better identify a subset of gastric cancers with poor prognosis than either L1 methylation or SAT-α methylation status alone. In the literature, there are two studies which investigated prognostic implications of SAT-α hypomethylation in human cancers, one in chronic lymphocytic leukemia [14] and the other in multiple myeloma [15]. However, no study is available regarding prognostic implications of SAT-α hypomethylation in solid cancers. To the best of our knowledge, the present study is the first to demonstrate the relationship between tumoral SAT-α hypomethylation and shortened survival in patients with gastric cancer. Further study is required to validate the relationship with another independent set of gastric cancers.
Although tumoral L1 hypomethylation has been demonstrated to be associated with shortened survival time in patients with gastric cancer, the mechanism of how tumoral L1 hypomethylation contributes to increased aggressiveness of gastric cancer remains unclear. Many researchers have suggested that diffuse genomic hypomethylation, represented by L1 hypomethylation, leads to chromosomal instability, which might contribute to increased aggressiveness of cancer cells. However, satellite hypomethylation is more likely to be related to chromosomal instability than L1 hypomethylation. Human cancer cells exhibit increased expression of satellite RNAs in association with diffuse genomic hypomethylation [16]. The demethylation of satellite sequences and expression of satellite RNAs might cause functional abnormalities in centromeres as the kinetochore in mitosis [5, 17]. In addition, the transfection of satellite sequences has been demonstrated to bring about chromosomal instability in murine cancer or human epithelial cell lines [5, 18].
In our current study, tumor areas with highest tumor density and representative histology were selected for manual dissection. However, the proportion of stromal and immune cells included in the dissected tumor areas varied case by case, and tumor purity ranged from 10 to 90% with a median of 50%. A concern might be raised over whether tumor purity affected the results of the survival analysis of L1 and SAT-α methylation statuses in the patients with advanced GC. To identify whether low L1 or SAT-α methylation status was a significant prognostic factor regardless of tumor purity, we divided gastric cancers into two subsets, gastric cancers with tumor purity < 50% (n = 231) and gastric cancers with tumor purity ≥ 50% (n = 243), and then performed survival analysis in the two subsets. Low methylation status of L1 or SAT-α was prognostic in the subset of tumor purity ≥ 50%, whereas in the subset of tumor purity < 50%, statistical significance was marginal in both OS and RFS (Supplementary Tables 5 and 6). These findings indicate that the relationship between the low methylation status of L1 or SAT-α and worse survival was more evident in gastric cancers with higher tumor purity.
L1 or SAT-α methylation level showed significant associations with prognostic parameters, including lymphatic invasion, venous invasion, and nodal stage, but did not exhibit significant associations with T stage or M stage. In contrast lymphatic or vascular invasion in which the presence of invasion was associated with a lower methylation level of L1, L1 methylation level tended to be higher in gastric cancers with perineural invasion than in those without perineural invasion. A similar trend was identified in a previous study in which L1 methylation levels were compared regarding their relationships with the clinicopathological features of intrahepatic cholangiocarcinomas; L1 methylation level was lower in tumors with lymphatic or venous invasion than in tumors without respective invasion but was not different between tumors with and without perineural invasion [19]. In Min et al.’s study analyzing the L1 methylation status in 22 gastric cancers, L1 methylation level was not different between gastric cancers with and without perineural invasion but was lower in gastric cancers with venous or lymphatic invasion than in gastric cancers without the respective invasion [20]. The pathogenesis of perineural invasion was traditionally hypothesized as tumor cells invading nerves because of their lowest resistance (due to loose and distensible perineural space) and thus spreading along nerves in an unopposed manner [21]. However, as evidenced by the present study in which 45% of advanced gastric cancers did not harbor perineural invasion, a considerable proportion of gastric cancers invade into proper muscle and subserosal layers without the involvement of nerves. Now, it is understood that perineural invasion is not a passive extension, but an active invasion facilitated by the perineural microenvironment, which secretes specific molecular signals, including growth factors and chemokines [22]. At present, it is unclear why tumors with perineural invasion tend to harbor higher L1 methylation levels than in tumors with no perineural invasion. Different molecular mechanisms are thought to operate between lymphatic or venous invasion and perineural invasion based on their contrasting associations with tumoral L1 methylation levels between lymphatic or venous invasion and perineural invasion.
Our study has demonstrated a close association between tumor histology (by Lauren classification) and methylation levels of L1 or SAT-α. Of the three histological types (except for the unclassified type), gastric cancers of the diffuse type showed the highest methylation level of L1 or SAT-α, whereas gastric cancers of the intestinal type showed the lowest methylation level of L1 or SAT-α, which is regardless of sex (Supplementary Table 7). Considering the fact that genome-wide DNA demethylation correlates with chromosomal instability in gastrointestinal tract carcinomas [23, 24] and that gastric cancers of the diffuse and intestinal type are genomically stable and unstable, respectively [25], methylation levels of repetitive DNA elements are expected to be higher in gastric cancers of the diffuse type than in gastric cancers of the intestinal type. Furthermore, difference in the frequency of TP53 mutations between histological types might contribute to the difference in the methylation levels of L1 retrotranspons between histological types because TP53 protein acts to restrain L1 activity and mutant TP53 proteins are disabled for this function [25, 26]. The significant difference of L1 or SAT-α methylation level between tumor subsites seems to be related to the significant difference in the proportion of the diffuse type between gastric cancers involving and not involving cardia (data not shown). The higher proportion of the diffuse type in gastric cancers involving cardia is thought to lead to higher methylation levels of L1 or SAT-alpha in gastric cancers involving cardia than in gastric cancers not involving cardia. Although aging-dependent DNA hypomethylation has been demonstrated in gastric cancers [24, 27], we found that aging-related DNA hypomethylation was not shared between male and female patients and among gastric cancers of the different histological types (Supplementary Figs. 1 and 2). For male patients, gastric cancers of the intestinal type or mixed type, except for the diffuse type, showed aging-related DNA hypomethylation, whereas gastric cancers from female patients did not exhibit, regardless of the histological type, aging-related DNA hypomethylation. The reason why age-associated difference in methylation levels of L1 or SAT-α was found in male patients, but not in female patients could be explained as follows: for gastric cancers from male patients, a significant difference existed in the proportion of the intestinal type between younger and older patients (36 vs. 53%) but there was no significant difference in the proportion of intestinal type for gastric cancers from female patients between younger and older group (15 vs. 31%). For male patients, methylation levels of L1 or SAT-α significantly decreased from younger group to older group because half of the gastric cancers from older patients was comprised by the intestinal type which showed aging-related hypomethylation of L1 or SAT-α. In contrast, for female patients, there was no significant increase in the proportion of the intestinal type with aging and the intestinal type did not show aging-related DNA hypomethylation.
Tournier et al. cast doubt upon the feasibility of pyrosequencing methylation assays in formalin-fixed, paraffin-embedded (FFPE) tissue samples as they demonstrated discrepancies in the measured methylation level of interrogated CpG sites between paired fresh-frozen and FFPE tissue samples [13]. In a previous study in which we measured methylation levels in four serial CpG sites of L1 using a pyrosequencing assay, differences in the measured value of L1 methylation levels between paired fresh-frozen and FFPE tissue samples existed, and the difference varied depending on CpG sites [11]. We thought that an artificial increase in the measured value of L1 methylation level might be related to the resistance of archival tissue DNA samples against thermal and alkaline denaturation because of the inter-strand DNA crosslinking and protein to DNA crosslinking caused by formalin fixation. However, we found that treating DNA samples with prolonged heating before bisulfite conversion decreased the difference in the measured value of L1 methylation levels between paired fresh-frozen and FFPE tissue samples. In the present study, we applied prolonged heat treatment to archival tissue DNA samples before bisulfite conversion and then performed a pyrosequencing methylation assay for both L1 and SAT-α.
In our study, overall survival was defined as time to death regardless of cause. RFS was defined as time to progression of disease, local recurrence, distant metastasis, or death, whichever comes first. Only reappearance of gastric cancer (but not non-gastric cancer) were defined as event for progress of disease, recurrence, and metastasis. Mean survival for RFS and OS did not show a vast difference in the current study. However, the discrepancy between OS and RFS outcomes are considered organ specific because the mean survival between OS and other survival outcomes could show a much smaller gap in gastric cancer than in other cancers such as adrenocortical carcinoma or testicular germ cell tumor [28]. In the current study, most gastric cancer patients with recurrence died within a short span of time, which might contribute to the smaller gap between OS and RFS. Besides, stage IV cases were included and comprised 12% of the cases, which also contributed to the overlap of RFS and OS.
In conclusion, we analyzed a consecutive series of 492 cases of advanced gastric carcinoma for their methylation status in L1 and SAT-α using a pyrosequencing methylation assay and found that both L1 and SAT-α hypomethylation statuses were independent prognostic parameters heralding poor prognosis in both OS and RFS. SAT-α methylation status can be used to identify a subset of gastric cancers with worse prognosis in gastric cancers with L1 hypomethylation.
References
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. https://doi.org/10.1038/nrg2640.
Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272(12):7810–6.
Montoya-Durango DE, Ramos KS. L1 retrotransposon and retinoblastoma: molecular linkages between epigenetics and cancer. Curr Mol Med. 2010;10(5):511–21.
Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, et al. Transcription of satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36(2):423–34.
Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103(23):8709–14.
Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–36.
Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219(4):410–6.
Bae JM, Shin SH, Kwon HJ, Park SY, Kook MC, Kim YW, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131(6):1323–31.
Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16(4):480–7.
Song YS, Kim Y, Cho NY, Yang HK, Kim WH, Kang GH. Methylation status of long interspersed element-1 in advanced gastric cancer and its prognostic implication. Gastric Cancer. 2016;19(1):98–106.
Wen X, Jeong S, Kim Y, Bae JM, Cho NY, Kim JH, et al. Improved results of LINE-1 methylation analysis in formalin-fixed, paraffin-embedded tissues with the application of a heating step during the DNA extraction process. Clin Epigenet. 2017;9:1.
Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125(3):723–9.
Tournier B, Chapusot C, Courcet E, Martin L, Lepage C, Faivre J, et al. Why do results conflict regarding the prognostic value of the methylation status in colon cancers? The role of the preservation method. BMC Cancer. 2012;12:12.
Fabris S, Bollati V, Agnelli L, Morabito F, Motta V, Cutrona G, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics. 2011;6(2):188–94.
Aoki Y, Nojima M, Suzuki H, Yasui H, Maruyama R, Yamamoto E, et al. Genomic vulnerability to LINE-1 hypomethylation is a potential determinant of the clinicogenetic features of multiple myeloma. Genome Med. 2012;4(12):101.
Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–6.
Rosic S, Kohler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol. 2014;207(3):335–49.
Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–84.
Jeong S, Lee K, Wen X, Kim Y, Cho NY, Jang JJ, et al. Tumoral LINE-1 hypomethylation is associated with poor survival of patients with intrahepatic cholangiocarcinoma. BMC Cancer. 2017;17(1):588.
Min J, Choi B, Han TS, Lee HJ, Kong SH, Suh YS, et al. Methylation levels of LINE-1 as a useful marker for venous invasion in both FFPE and frozen tumor tissues of gastric cancer. Mol Cells. 2017;40(5):346–54.
Batsakis JG. Nerves and neurotropic carcinomas. Ann Otol Rhinol Laryngol. 1985;94(4 Pt 1):426–7.
Amit M, Na’ara S, Gil Z. Mechanisms of cancer dissemination along nerves. Nat Rev Cancer. 2016;16(6):399–408.
Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66(17):8462–9468.
Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9(3):199–207.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9.
Wylie A, Jones AE, D’Brot A, Lu WJ, Kurtz P, Moran JV, et al. p53 genes function to restrain mobile elements. Genes Dev. 2016;30(1):64–77.
Saito M, Suzuki K, Maeda T, Kato T, Kamiyama H, Koizumi K, et al. The accumulation of DNA demethylation in Sat alpha in normal gastric tissues with Helicobacter pylori infection renders susceptibility to gastric cancer in some individuals. Oncol Rep. 2012;27(6):1717–25.
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–16.
Acknowledgements
This work was supported by a grant from the Basic Science Research Program through the National Research Foundation (NRF) funded by the Korea Ministry of Science, ICT and Future Planning (2011-0030049; 2016M3A9B6026921) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Korean Ministry of Health and Welfare (HI14C1277).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
This study was approved by Seoul National University Hospital Institute Review of Board, which waived the requirement to obtain informed consent (IRB No. 1312-051-542) and was conducted in accordance with the Declaration of Helsinki.
Conflict of interest
The authors have declared that no competing interests exists.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10120_2018_852_MOESM8_ESM.tif
Supplementary Figure 1. Correlation between L1 methylation level and age according to histological type and sex (TIF 414 KB)
10120_2018_852_MOESM9_ESM.tif
Supplementary Figure 2. Correlation between SAT-α methylation level and age according to histological type and sex (TIF 434 KB)
Rights and permissions
About this article
Cite this article
Kim, Y., Wen, X., Jeong, S. et al. Combinatory low methylation statuses of SAT-α and L1 are associated with shortened survival time in patients with advanced gastric cancer. Gastric Cancer 22, 37–47 (2019). https://doi.org/10.1007/s10120-018-0852-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-018-0852-8