Abstract
Purpose
Rapid, reliable identification of mycobacteria from positive cultures is essential for patient management, particularly for the differential diagnosis of Mycobacterium tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM) species. The aim of the present study was to evaluate a new “In-Vitro-Diagnostic”-certified PCR kit, FluoroType®-Mycobacteria VER 1.0 (Hain Lifescience GmbH) for NTM and MTBC identification from cultures.
Methods
Mycobacteria identification isolated from positive cultures during routine practice at the Lyon university hospital mycobacteria laboratory obtained by hsp65 amplification/sequencing were compared retrospectively and prospectively to those obtained by and the FluoroType®-Mycobacteria VER 1.0 kit.
Results
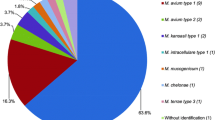
The overall agreement between hsp65 amplification/sequencing and the FluoroType®-Mycobacteria VER 1.0 kit was 88.4% (84/95); 91.2% (52/57) for the retrospective period and 84.2% (32/38) for the prospective period. There were 9 (9.5%) minor discrepancies (species in the FluoroType®-Mycobacteria VER 1.0 database and identified at genus level): 4 during the retrospective period, 5 during the prospective period; and 2 (2.1%) major discrepancies (species in the FluoroType®-Mycobacteria VER 1.0 database and identified incorrectly to species level): 1 during the retrospective period (M. kumamotonense identified as M. abscessus subsp massiliense by the kit) and 1 during the prospective period (M. chimaera identified as M. smegmatis by the kit). Including concordant results at genus level and minor discrepancies, 17.9% (17/95) of strains were identified as Mycobacterium sp. by the FluoroType®-Mycobacteria-VER 1.0 kit.
Conclusion
The good performance of the FluoroType®-Mycobacteria-VER 1.0 kit with few major discrepancies could enable its use for first-line identification of positive mycobacteria cultures. However, an alternative identification method at least for reference laboratories is needed owing to the non-negligible proportion of NTM strains were identified at genus level.
Similar content being viewed by others
Data availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Global tuberculosis report 2022 [Internet]. [cited 2023 29];doi: https://www.who.int/publications-detail-redirect/9789240061729
Dahl VN, Mølhave M, Fløe A, van Ingen J, Schön T, Lillebaek T et al (2022) Global trends of pulmonary infections with nontuberculous mycobacteria: a systematic review. Int J Infect Dis IJID off Publ Int Soc Infect Dis 125:120–131. https://doi.org/10.1016/j.ijid.2022.10.013
Koh W-J, Nontuberculous, Mycobacteria—Overview (2017) 27;5 Microbiol. Spectr. :https://doi.org/10.1128/microbiolspec.tnmi7-0024-2016. doi: 10.1128/microbiolspec.tnmi7-0024-2016
Forbes BA, Hall GS, Miller MB, Novak SM, Rowlinson M-C, Salfinger M et al (2018) Practice guidelines for clinical Microbiology Laboratories: Mycobacteria. Clin Microbiol Rev 31. https://doi.org/10.1128/CMR.00038-17
Huh HJ, Kim S-Y, Shim HJ, Kim DH, Yoo IY, Kang O-K et al (2019) GenoType NTM-DR performance evaluation for identification of Mycobacterium avium Complex and Mycobacterium abscessus and determination of Clarithromycin and Amikacin Resistance. J Clin Microbiol 57:e00516–e00519. https://doi.org/10.1128/JCM.00516-19
Kehrmann J, Kurt N, Rueger K, Bange F-C, Buer J (2016) GenoType NTM-DR for identifying Mycobacterium abscessus subspecies and determining Molecular Resistance. J Clin Microbiol 54:1653–1655. https://doi.org/10.1128/JCM.00147-16
Mäkinen J, Marjamäki M, Marttila H, Soini H (2006) Evaluation of a novel strip test, GenoType Mycobacterium CM/AS, for species identification of mycobacterial cultures. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Dis 12:481–483. https://doi.org/10.1111/j.1469-0691.2006.01380.x
Lee AS, Jelfs P, Sintchenko V, Gilbert GL (2009) Identification of non-tuberculous mycobacteria: utility of the GenoType Mycobacterium CM/AS assay compared with HPLC and 16S rRNA gene sequencing. J Med Microbiol 58:900–904. https://doi.org/10.1099/jmm.0.007484-0
Pastrone L, Curtoni A, Criscione G, Scaiola F, Bottino P, Guarrasi L et al (2023) Evaluation of two different Preparation protocols for MALDI-TOF MS nontuberculous mycobacteria identification from Liquid and Solid Media. Microorganisms 3:11:120. https://doi.org/10.3390/microorganisms11010120
Rodríguez-Temporal D, Zvezdánova ME, Benedí P, Marín M, Blázquez-Sánchez M, Ruiz-Serrano MJ et al (2022) 20;doi Identification of Nocardia and non-tuberculous Mycobacterium species by MALDI-TOF MS using the VITEK MS coupled to IVD and RUO databases. Microb. Biotechnol. : https://doi.org/10.1111/1751-7915.14146
Rindi L, Puglisi V, Franconi I, Fais R, Lupetti A Rapid and Accurate Identification of Nontuberculous Mycobacteria directly from positive primary MGIT cultures by MALDI-TOF MS. Microorganisms 2022 18;10:1447. https://doi.org/10.3390/microorganisms10071447
Martin EC, Limousin L, Renaux C, Catherinot E, Vasse M (2023) Evaluation of the mycobacteria MBT kit for identification of nontuberculous mycobacteria by MALDI-TOF biotyper (Bruker). Diagn. Microbiol Infect Dis 107:116044. https://doi.org/10.1016/j.diagmicrobio.2023.116044
Xu Y, Liang B, Du C, Tian X, Cai X, Hou Y et al (2019) Rapid Identification of clinically relevant Mycobacterium species by Multicolor Melting curve analysis. J Clin Microbiol 2. https://doi.org/10.1128/jcm.01096-18
Ratnam S, Stead FA, Howes M (1987) Simplified acetylcysteine-alkali digestion-decontamination procedure for isolation of mycobacteria from clinical specimens. J Clin Microbiol 25:1428–1432
Genestet C, Baffert Y, Vallée M, Bernard A, Benito Y, Lina G et al (2022) Development, evaluation, and implementation of a House-made targeted next-generation sequencing spoligotyping in a French laboratory. Int J Mol Sci 25:23:11302. https://doi.org/10.3390/ijms231911302
Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T (1993) Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 31:175–178. https://doi.org/10.1128/jcm.31.2.175-178.1993
Devulder G, Perrière G, Baty F, Flandrois JP (2003) BIBI, a Bioinformatics Bacterial Identification Tool. J Clin Microbiol 41:1785–1787. https://doi.org/10.1128/JCM.41.4.1785-1787.2003
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159–174
Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F et al (2007) An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 15:175:367–416. https://doi.org/10.1164/rccm.200604-571ST
Clinical and laboratory standards institute (2018) M62 - Performance Standards for Susceptibility testing of Mycobacetria, Nocardia spp., and other aerobic actinomycetes. 1St Edition
Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C et al (2020) Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J 56:2000535. https://doi.org/10.1183/13993003.00535-2020
Tortoli E, Kohl TA, Brown-Elliott BA, Trovato A, Leão SC, Garcia MJ et al (2016) Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. Abscessus and Mycobacteriumabscessus subsp. bolletii and designation of Mycobacteriumabscessus subsp. massiliense comb. Nov. Int J Syst Evol Microbiol 66:4471–4479. https://doi.org/10.1099/ijsem.0.001376
Dippenaar A, Derendinger B, Dolby T, Beylis N, van Helden PD, Theron G et al (2021) Diagnostic accuracy of the FluoroType MTB and MTBDR VER 2.0 assays for the centralized high-throughput detection of Mycobacterium tuberculosis complex DNA and isoniazid and rifampicin resistance. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Dis 27:1351e1–1351e4. https://doi.org/10.1016/j.cmi.2021.04.022
Acknowledgements
The authors thank Philip Robinson (DRS, Hospices Civils de Lyon, Lyon, France) for help with manuscript preparation.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Lisa Piasecki, Charlotte Genestet, Yvonne Benito, Jean-Philippe Rasigade, Gérard Lina, Oana Dumitrescu and Elisabeth Hodille. The first draft of the manuscript was written by Elisabeth Hodille and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and informed consent
The present study was in accordance with the decision of the ethics committee of the Lyon university hospital, France (declared sample collection: DC-2011-1306). In accordance with French legislation, written informed patient consent was not required to compare the technical performance of assays with clinical specimens or clinical isolates collected following clinical recommendations.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Piasecki, L., Genestet, C., Benito, Y. et al. Retrospective and prospective evaluation of the FluoroType®-Mycobacteria VER 1.0 assay for the identification of mycobacteria from cultures in a French center. Eur J Clin Microbiol Infect Dis (2024). https://doi.org/10.1007/s10096-024-04825-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10096-024-04825-8