Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV‐2) caused the coronavirus disease 2019 (COVID-19), leading to a global pandemic. The molecular diagnosis of this virus is mostly performed by collecting upper respiratory samples, which has many disadvantages, including patient discomfort and the need for trained healthcare professionals. Although saliva has emerged as a more comfortable sample, the use of additives to preserve viral RNA is expensive and, in some cases, difficult for self-collection.
Method
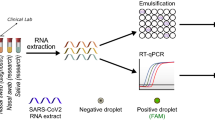
This study evaluated the diagnostic performance by RT-PCR and stability of self-collected saliva using wide-mouth specimen collection cups without stabilization and/or inactivation buffers for SARS-CoV-2 detection, compared to nasopharyngeal samples and saliva collected with additives. Additionally, the study assessed the acceptability of this sample collection method among participants and healthcare personnel.
Results
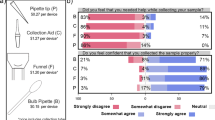
The study included 1281 volunteers with a 24.6% positive infection rate. Saliva demonstrated comparable diagnostic performance to nasopharyngeal samples, with a sensitivity of 87.6% and specificity of 99.6%, for a total percent agreement of 96.4%. The study also showed that viral RNA in saliva remained stable for at least 72 h at different temperatures. Notably, saliva samples without additives exhibited a lower RdRp Ct compared to samples with additives, suggesting that the absence of stabilization and/or inactivation buffers does not significantly affect its performance. The study highlighted the acceptability of saliva among patients and healthcare personnel due to its noninvasive nature and ease of collection.
Conclusions
This research supports the implementation of self-collected saliva as a comfortable and user-friendly alternative sample for SARS-CoV-2 diagnosis.




Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
World Health Organization WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [Internet]. 2023 [cited 2023 Feb 15]. Available from: https://covid19.who.int/
Liu BM, Hill HR (2020) Role of host Immune and inflammatory responses in COVID-19 cases with underlying primary immunodeficiency: a review. J Interferon Cytokine Res 40:549–554
Liu BM, Martins TB, Peterson LK, Hill HR (2021) Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: a review. Cytokine 142:155478
Kandel CE, Young M, Serbanescu MA, Powis JE, Bulir D, Callahan J et al (2021) Detection of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) in outpatients: a multicenter comparison of self-collected saline gargle, oral swab, and combined oral-anterior nasal swab to a provider collected nasopharyngeal swab. Infect Control Hosp Epidemiol 42:1340–1344
Moisset X, Gautier N, Godet T, Parabère S, Pereira B, Meunier E et al (2021) Nasopharyngeal swab-induced pain for SARS-CoV-2 screening: a randomised controlled trial of conventional and self-swabbing. Eur J Pain 25:924–929
Marra P, Colacurcio V, Bisogno A, de Luca P, Calvanese M, Petrosino M et al (2021) Evaluation of discomfort in nasopharyngeal swab specimen collection for SARS-CoV-2 diagnosis. Clin Ter 172:448–452
Deressa W, Worku A, Abebe W, Gizaw M, Amogne W (2021) Risk perceptions and preventive practices of COVID-19 among healthcare professionals in public hospitals in Addis Ababa, Ethiopia. PLoS ONE 16:e0242471
Callahan C, Ditelberg S, Dutta S, Littlehale N, Cheng A, Kupczewski K et al (2021) Saliva is comparable to nasopharyngeal swabs for Molecular Detection of SARS-CoV-2. Microbiol Spectr. ;9
Felix AC, de Paula AV, Ribeiro AC, da Silva FC, Inemami M, Costa AA et al (2022) Saliva as a reliable sample for COVID-19 diagnosis in paediatric patients. Int J Paediatr Dent 32:123–125
Duncan DB, Mackett K, Ali MU, Yamamura D, Balion C (2023) Performance of saliva compared with nasopharyngeal swab for diagnosis of COVID-19 by NAAT in cross-sectional studies: systematic review and meta-analysis. Clin Biochem 117:84–93
Allicock OM, Petrone ME, Yolda-Carr D, Breban M, Walsh H, Watkins AE et al (2022) Evaluation of saliva self-collection devices for SARS-CoV-2 diagnostics. BMC Infect Dis. ;22
Gutierrez Hincapie S, Salazar Herrera R, Álvarez Acevedo LC, DETECCIÓN DE SARS-COV-2 POR, RT-QPCR, EN MUESTRAS DE SALIVA Ayudas Diagnósticas SURA [Internet], Medellín C (2021) [cited 2024 Mar 4]. Available from: https://www.ins.gov.co/BibliotecaDigital/Deteccion-SARS-COV%202-PCR-saliva-SURA.pdf
Minsalud Lineamientos para el uso de pruebas moleculares RT-PCR, pruebas de antígeno y pruebas serológicas para SARS-CoV-2 (COVID-19) en Colombia [Internet]. 2020 [cited 2022 May 16]. p. 1–20. Available from: https://acin.org/images/guias/coronavirus/GIPS21_LINEAMIENTO_DE-PRUEBAS_22_07_2020.pdf
BIONEER COVID-19_COVID-19 Real-Time RT-PCR Kit_BIONEER [Internet]. 2020 [cited 2023 Sep 20]. Available from: https://eng.bioneer.com/20-ncv-1111.html
INS COVID-19 en Colombia [Internet]. 2023 [cited 2023 Jul 6]. Available from: https://www.ins.gov.co/Noticias/Paginas/coronavirus-departamento.aspx
Genelhoud G, Adamoski D, Spalanzani RN, Bochnia-Bueno L, de Oliveira JC, Gradia DF et al (2022) Comparison of SARS-CoV-2 molecular detection in nasopharyngeal swab, saliva, and gargle samples. Diagn Microbiol Infect Dis 103:115678
Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG et al (2021) Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and Meta-analysis. JAMA Intern Med 181:353–360
INS. Protocolo de verificación (validación secundaria) para pruebas moleculares de PCR en tiempo real (RT-qPCR) para la detección del SARS-CoV-2. Dirección Redes en Salud Pública Dirección Vigilancia y Análisis del Riesgo [Internet] (2021) Available from: http://www.saludcapital.gov.co/CTDLab/Publicaciones/2021/Protocolo_verifi_RT_PCR_SARS_CoV-2.pdf
Beyene GT, Alemu F, Kebede ES, Alemayehu DH, Seyoum T, Tefera DA et al (2021) 11:1 Saliva is superior over nasopharyngeal swab for detecting SARS-CoV2 in COVID-19 patients. Scientific Reports 2021;11:22640
Jung EJ, Lee SK, Shin SH, Kim JS, Woo H, Cho EJ et al (2023) Comparison of nasal swabs, nasopharyngeal swabs, and Saliva samples for the detection of SARS-CoV-2 and other respiratory virus infections. Ann Lab Med 43:434–442
Winnett AV, Akana R, Shelby N, Davich H, Caldera S, Yamada T et al (2023) Extreme differences in SARS-CoV-2 viral loads among respiratory specimen types during presumed pre-infectious and infectious periods. PNAS Nexus 2:1–16
Griesemer SB, Van Slyke G, Ehrbar D, Strle K, Yildirim T, Centurioni DA et al (2021) Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. J Clin Microbiol 59:e01418–e01420
Ott IM, Strine MS, Watkins AE, Boot M, Kalinich CC, Harden CA et al (2020) Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv 27:1146–1150
Perret PC, Abarca K, Solari S, Aguilera P, García-Huidobro D, Olivares D et al (2022) Detección Del SARS-CoV-2 mediante RT-qPCR utilizando saliva en pacientes ambulatorios con estudio de COVID-19. Rev Chil Infectol 39:372–381
Nacher M, Mergeay-Fabre M, Blanchet D, Benois O, Pozl T, Mesphoule P et al (2021) Diagnostic accuracy and acceptability of molecular diagnosis of COVID-19 on saliva samples relative to nasopharyngeal swabs in tropical hospital and extra-hospital contexts: the COVISAL study. PLoS ONE 16:e0257169
McLennan K, Barton E, Lang C, Adams IR, McAllister G, Reijns MAM et al (2022) User acceptability of saliva and gargle samples for identifying COVID-19 positive high-risk workers and household contacts. Diagn Microbiol Infect Dis 104:115732
To KKW, Yip CCY, Lai CYW, Wong CKH, Ho DTY, Pang PKP et al (2019) Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect 25:372–378
Laxton CS, Peno C, Hahn AM, Allicock OM, Perniciaro S, Wyllie AL (2023) The potential of saliva as an accessible and sensitive sample type for the detection of respiratory pathogens and host immunity. Lancet Microbe 4:e837–e850
Liu B, Totten M, Nematollahi S, Datta K, Memon W, Marimuthu S et al (2020) Development and evaluation of a fully automated Molecular Assay Targeting the mitochondrial small subunit rRNA gene for the detection of Pneumocystis Jirovecii in Bronchoalveolar Lavage fluid specimens. J Mol Diagn 22:1482–1493
Sueki A, Matsuda K, Yamaguchi A, Uehara M, Sugano M, Uehara T et al (2016) Evaluation of saliva as diagnostic materials for influenza virus infection by PCR-based assays. Clin Chim Acta 453:71–74
To KK, Lu L, Yip CC, Poon RW, Fung AM, Cheng A et al (2017) Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect 6:e49
Acknowledgements
We express our gratitude to Ayudas Diagnósticas SURA, as sponsoring and executing entity. We extend our heartfelt thanks to all the volunteers who willingly participated in the study. We would like to acknowledge the dedication and efforts of the healthcare personnel responsible for recruiting patients and collecting samples, as well as the molecular biology professionals who processed the collected samples. To the Biosciences management, and its research and development team, as well as the SURA personalized Medicine research group, for their unwavering support in implementing recruitment strategies. To all those involved, their contributions have been instrumental in the successful completion of this research.
Funding
This study was funded by Ayudas Diagnósticas SURA-Colombia.
Author information
Authors and Affiliations
Contributions
Conceptualization: H del BA, S. G.-H, L.A.-A and C.M.-E. Methodology: L.P.-Z., S. G.-H., L.A.-A., Y.G.-C, C.M.-E., C.P.-O, J.B.-M. Statistical analysis: L.P.-Z. and C.M.-E. Drafting of the manuscript: C.M.-E. Analysis, or interpretation of data: L.P.-Z., S. G.-H., L.A.-A., Y.G.-C, C.M.-E., C.P.-O, Y.G.-C, M.A.-P, D.T.-D., J.B.-M., M.-G.-G. All authors reviewed and approved the final version.
Corresponding author
Ethics declarations
Financial interest
The authors declare no financial interests.
Ethics approval
The Committee of Ethics and Good Clinical Practices in Health Research (CEI-SURA) approved the study (Act number 62).
Patient consent
All volunteers who participated in the study signed an informed consent. For children who agreed to participate, the informed consent was signed by their father/mother or legal representative.
Conflict of interest
The authors declare no conflicts of interest with this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marín-Echeverri, C., Pérez-Zapata, L., Álvarez-Acevedo, L. et al. Diagnostic performance, stability, and acceptability of self-collected saliva without additives for SARS-CoV-2 molecular diagnosis. Eur J Clin Microbiol Infect Dis (2024). https://doi.org/10.1007/s10096-024-04819-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10096-024-04819-6