Abstract
Purpose
Population-based surveillance was undertaken to determine clinical factors, susceptibility patterns, and incidence rates (IR) of Pseudomonas aeruginosa causing bloodstream infections (BSIs) in a Canadian region (2010–2018).
Methods
We combined clinical data with genomics to characterize P. aeruginosa (BSIs) (n = 167) in a well-defined Canadian (Calgary) human population over a 9-year period (2010–2018).
Results
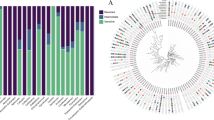
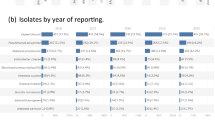
The annual population IR per 100,000 patient years increased from 3.4/100,000 in 2010 to 5.9/100,000 in 2018, with the highest IRs in elderly males from the hospital setting. Over a quarter of patients presented with febrile neutropenia, followed by urinary tract infections and pneumonia. Antimicrobial resistance (AMR) rates and determinants were rare. The P. aeruginosa population was polyclonal consisting of three dominant sequence types (STs), namely ST244, ST111, and ST17. Antimicrobial-susceptible ST244 was the most common clone and belonged to three clades (A, B, C). The ST244 IR/100,000 increased over time due to the expansion of clade C. Multidrug-resistant ST111 was the second most common clone and IR/100,000 decreased over time. ST111 belonged to three clades (A, B, C) with clade C containing blaVIM-2. Different serotypes were linked to various STs. The IR/100,000 of P. aeruginosa that belonged to serotypes O6 increased significantly over time.
Conclusion
An effective multivalent vaccine consisting of five serotypes (O1, O3, O5, O6, O11) would confer protection to > 70% of Calgary residents with P. aeruginosa BSIs. This study has provided a unique perspective of the population dynamics over time of P. aeruginosa STs, clades, and serotypes responsible for BSIs.


Similar content being viewed by others
Data availability
Sequence data was uploaded to NCBI (BioProject PRJNA988909).
Code availability
Sequence data is available at NCBI (BioProject PRJNA988909).
References
Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, Jones RN (2019) The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 63(7):e00355–19. https://doi.org/10.1128/AAC.00355-19
Holmes CL, Anderson MT, Mobley HLT, Bachman MA (2021) Pathogenesis of gram-negative bacteremia. Clin Microbiol Rev 34(2):e00234-20. https://doi.org/10.1128/CMR.00234-20
Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging infections program healthcare-associated I, antimicrobial use prevalence survey T (2014) Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370(13):1198-1208
Kang CI, Kim SH, Kim HB, Park SW, Choe YJ, Oh MD, Kim EC, Choe KW (2003) Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 37(6):745–751
Kontula KS, Skogberg K, Ollgren J, Jarvinen A, Lyytikainen O (2022) Early deaths associated with community-acquired and healthcare-associated bloodstream infections: a population-based study, Finland, 2004 to 2018. Euro Surveill 27(36):2101067. https://doi.org/10.2807/1560-7917.ES.2022.27.36.2101067
Laupland KB (2013) Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect 19(6):492–500
Holland MS, Nobrega D, Peirano G, Naugler C, Church DL, Pitout JDD (2020) Molecular epidemiology of Escherichia coli causing bloodstream infections in a centralized Canadian region: a population-based surveillance study. Clin Microbiol Infect 26(11):1554 e1551-1554 e1558
Laupland KB, Gregson DB, Church DL, Ross T, Pitout JD (2008) Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clin Microbiol Infect 14(11):1041–1047
Stokes W, Peirano G, Matsumara Y, Nobrega D, Pitout JDD (2022) Population-based surveillance of Enterobacter cloacae complex causing blood stream infections in a centralized Canadian region. Eur J Clin Microbiol Infect Dis 41(1):119–125
Peirano G, Lynch T, Matsumara Y, Nobrega D, Finn TJ, DeVinney R, Pitout JDD (2020) Trends in population dynamics of Escherichia coli sequence type 131, Calgary, Alberta, Canada, 2006–2016(1). Emerg Infect Dis 26(12):2907–2915
Peirano G, Matsumara Y, Nobrega D, DeVinney R, Pitout J (2021) Population-based epidemiology of Escherichia coli ST1193 causing blood stream infections in a centralized Canadian region. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-021-04373-5
Pitout JDD (2021) Population dynamics of Escherichia coli causing bloodstream infections over extended time periods. mSphere 6(6):e0095621
Da Silva R, Casella T (2022) Healthcare-associated infections in patients who are immunosuppressed due to chemotherapy treatment: a narrative review. J Infect Dev Ctries 16(12):1784–1795
Sloot R, Nsonwu O, Chudasama D, Rooney G, Pearson C, Choi H, Mason E, Springer A, Gerver S, Brown C, Hope R (2022) Rising rates of hospital-onset Klebsiella spp. and Pseudomonas aeruginosa bacteraemia in NHS acute trusts in England: a review of national surveillance data, August 2020-February 2021. J Hosp Infect 119:175–181
Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ (2002) Health care–associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137(10):791–797
CLSI (2015) Performance standards for antimicrobial susceptibility testing; twenty-fifth information supplement. CLSI document M100-S25 (ISBN 1–56238–990–4). Clinical and Laboratory Standards Institute. 950 West Valley Road, Suit 2500, Wayne, Pennsylvania19087. USA
Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom KA, Ehlers MM, Mbelle NM, Shashkina E, Haslam DB, Dhawan P, Donnelly RJ, Chen L, Kreiswirth BN, Pitout JDD (2019) Klebsiella pneumoniae ST307 with bla(OXA-181,) South Africa, 2014–2016. Emerg Infect Dis 25(4):739–747
Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, Kreiswirth BN, Pitout JDD (2018) Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg Infect Dis 24(6):1010–1019
Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20(10):714–737
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Thrane SW, Taylor VL, Lund O, Lam JS, Jelsbak L (2016) Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J Clin Microbiol 54(7):1782–1788
Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Cheng L, Connor TR, Siren J, Aanensen DM, Corander J (2013) Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30(5):1224–1228
Nobrega D, Peirano G, Lynch T, Finn TJ, Devinney R, Pitout JDD (2021) Spatial distribution of Escherichia coli ST131 C subclades in a centralized Canadian urban region. J Antimicrob Chemother 76(5):1135–1139
Parkins MD, Gregson DB, Pitout JD, Ross T, Laupland KB (2010) Population-based study of the epidemiology and the risk factors for Pseudomonas aeruginosa bloodstream infection. Infection 38(1):25–32
Laupland KB, Parkins MD, Church DL, Gregson DB, Louie TJ, Conly JM, Elsayed S, Pitout JD (2005) Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-beta-lactamase (MBL)-producing strains. J Infect Dis 192(9):1606–1612
Parkins MD, Pitout JDD, Church DL, Conly JM, Laupland KB (2007) Treatment of infections caused by metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region. Clin Microbiol Infect 13(2):199–202
Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, Do T, Lanotte P, Mastouri M, Elghmati MS, Rojo F, Mejdi S, Giske CG (2011) Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS ONE 6(10):e25617
Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious D (2012) Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56(12):6349–6357
Recio R, Villa J, Viedma E, Orellana MA, Lora-Tamayo J, Chaves F (2018) Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents 52(2):172–179
Del Barrio-Tofino E, Lopez-Causape C, Oliver A (2020) Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired beta-lactamases: 2020 update. Int J Antimicrob Agents 56(6):106196
Sadek M, Le Guern R, Kipnis E, Gosset P, Poirel L, Dessein R, Nordmann P (2023) Progressive in vivo development of resistance to cefiderocol in Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 42(1):61–66
Pitout JD, Chow BL, Gregson DB, Laupland KB, Elsayed S, Church DL (2007) Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J Clin Microbiol 45(2):294–298
Thrane SW, Taylor VL, Freschi L, Kukavica-Ibrulj I, Boyle B, Laroche J, Pirnay JP, Levesque RC, Lam JS, Jelsbak L (2015) The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. mBio 6(5):e01396-01315
Stanislavsky ES, Lam JS (1997) Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol Rev 21(3):243–277
Nasrin S, Hegerle N, Sen S, Nkeze J, Sen S, Permala-Booth J, Choi M, Sinclair J, Tapia MD, Johnson JK, Sow SO, Thaden JT, Fowler VG Jr, Krogfelt KA, Brauner A, Protonotariou E, Christaki E, Shindo Y, Kwa AL, Shakoor S, Singh-Moodley A, Perovic O, Jacobs J, Lunguya O, Simon R, Cross AS, Tennant SM (2022) Distribution of serotypes and antibiotic resistance of invasive Pseudomonas aeruginosa in a multi-country collection. BMC Microbiol 22(1):13
Engel J, Balachandran P (2009) Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 12(1):61–66
Pena C, Cabot G, Gomez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodriguez-Bano J, Rodriguez-Lopez F, Tubau F, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious D (2015) Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60(4):539–548
Fischer S, Dethlefsen S, Klockgether J, Tummler B (2020) Phenotypic and genomic comparison of the two most common ExoU-positive pseudomonas aeruginosa clones, PA14 and ST235. mSystems 5(6):e01007-20. https://doi.org/10.1128/mSystems.01007-20
Funding
This work was supported by a research grant from the Alberta Precision Laboratories (#10026137).
Author information
Authors and Affiliations
Contributions
All authors designed the study and approved the manuscript. GP, YM, and DN performed WGS, bioinformatics, and statistical analysis. JP, DN, and DC combined the clinical and genomic data. JP wrote the first draft of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Ethics approval for this study was obtained through the University of Calgary Conjoint Health Research Ethics Board (REB18-1123).
Consent to participate
Not applicable.
Consent for publication
This manuscript has not been published and is not being considered for publication elsewhere.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peirano, G., Matsumara, Y., Nobrega, D. et al. Population-based genomic surveillance of Pseudomonas aeruginosa causing bloodstream infections in a large Canadian health region. Eur J Clin Microbiol Infect Dis 43, 501–510 (2024). https://doi.org/10.1007/s10096-024-04750-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-024-04750-w