Abstract
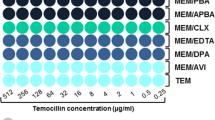
The objectives of this study were to define the shortest incubation times on the WASPLab for reliable MALDI-TOF/MS-based species identification and for the preparation of a 0.5 McFarland suspension for antimicrobial disk diffusion susceptibility testing using short subcultures growing on solid culture media inoculated by positive blood cultures spiked with a wide range of pathogens associated with bloodstream infections. The 520 clinical strains (20 × 26 different species) included in this study were obtained from a collection of non-consecutive and non-duplicate pathogens identified at Geneva University Hospitals. After 4 h of incubation on the WASPLab, microorganisms’ growth allowed accurate identification of 73% (380/520) (95% CI, 69.1–76.7%) of the strains included in this study. The identification rate increased to 85% (440/520) (95% CI, 81.3–87.5%) after 6-h incubation. When excluding Corynebacterium and Candida spp., the microbial growth was sufficient to permit accurate identification of all tested species (100%, 460/460) (95% CI, 99.2–100%) after 8-h incubation. With the exception of Burkholderia cepacia and Haemophilus influenzae, AST by disk diffusion could be performed for Enterobacterales and non-fermenting Gram-negative bacilli after only 4 h of growth in the WASPLab. The preparation of a 0.5 McFarland suspension for Gram-positive bacteria required incubation times ranging between 3 and 8 h according to the bacterial species. Only Corynebacterium spp. required incubation times as long as 16 h. The WASPLab enables rapid pathogen identification as well as swift comprehensive AST from positive blood cultures that can be implemented without additional costs nor hands-on time by defining optimal time points for image acquisition.




Similar content being viewed by others
References
Gaieski DF, Mikkelsen ME, Band RA, Pines JM, Massone R, Furia FF, Shofer FS, Goyal M (2010) Impact of time to antibiotics on survival in patients with severe sepsis or septic shock in whom early goal-directed therapy was initiated in the emergency department. Crit Care Med 38(4):1045–1053
Larche J, Azoulay E, Fieux F, Mesnard L, Moreau D, Thiery G, Darmon M, Le Gall JR, Schlemmer B (2003) Improved survival of critically ill cancer patients with septic shock. Intensive Care Med 29(10):1688–1695
Garnacho-Montero J, Aldabo-Pallas T, Garnacho-Montero C, Cayuela A, Jimenez R, Barroso S, Ortiz-Leyba C (2006) Timing of adequate antibiotic therapy is a greater determinant of outcome than are TNF and IL-10 polymorphisms in patients with sepsis. Crit Care 10(4):R111
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6):1589–1596
Leal HF, Azevedo J, Silva GEO, Amorim AML, de Roma LRC, Arraes ACP, Gouveia EL, Reis MG, Mendes AV, de Oliveira SM, Barberino MG, Martins IS, Reis JN (2019) Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis 19(1):609
Matos ECO, Andriolo RB, Rodrigues YC, Lima PDL, Carneiro I, Lima KVB (2018) Mortality in patients with multidrug-resistant Pseudomonas aeruginosa infections: a meta-analysis. Rev Soc Bras Med Trop 51(4):415–420
Liesenfeld O, Lehman L, Hunfeld KP, Kost G (2014) Molecular diagnosis of sepsis: new aspects and recent developments. Eur J Microbiol Immunol (Bp) 4(1):1–25
Candel FJ, Borges Sa M, Belda S, Bou G, Del Pozo JL, Estrada O, Ferrer R, Gonzalez Del Castillo J, Julian-Jimenez A, Martin-Loeches I, Maseda E, Matesanz M, Ramirez P, Ramos JT, Rello J, Suberviola B, Suarez de la Rica A, Vidal P (2018) Current aspects in sepsis approach. Turning things around. Rev Esp Quimioter 31(4):298–315
Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J (2010) Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol 48(4):1169–1175
Ferreira L, Sanchez-Juanes F, Porras-Guerra I, Garcia-Garcia MI, Garcia-Sanchez JE, Gonzalez-Buitrago JM, Munoz-Bellido JL (2011) Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect 17(4):546–551
Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M (2010) Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol 48(5):1584–1591
Haigh JD, Green IM, Ball D, Eydmann M, Millar M, Wilks M (2013) Rapid identification of bacteria from bioMerieux BacT/ALERT blood culture bottles by MALDI-TOF MS. Br J Biomed Sci 70(4):149–155
Campigotto A, Goneau L, Matukas LM (2018) Direct identification and antimicrobial susceptibility testing of microorganisms from positive blood cultures following isolation by lysis-centrifugation. Diagn Microbiol Infect Dis 92(3):189–193
Meex C, Neuville F, Descy J, Huynen P, Hayette MP, De Mol P, Melin P (2012) Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J Med Microbiol 61(Pt 11):1511–1516
Klein S, Zimmermann S, Kohler C, Mischnik A, Alle W, Bode KA (2012) Integration of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in blood culture diagnostics: a fast and effective approach. J Med Microbiol 61(Pt 3):323–331
Kohlmann R, Hoffmann A, Geis G, Gatermann S (2015) MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. Int J Med Microbiol 305(4–5):469–479
Thomin J, Aubin GG, Foubert F, Corvec S (2015) Assessment of four protocols for rapid bacterial identification from positive blood culture pellets by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Vitek(R) MS). J Microbiol Methods 115:54–56
March GA, Garcia-Loygorri MC, Simarro M, Gutierrez MP, Orduna A, Bratos MA (2015) A new approach to determine the susceptibility of bacteria to antibiotics directly from positive blood culture bottles in two hours. J Microbiol Methods 109:49–55
Jakovljev A, Bergh K (2015) Development of a rapid and simplified protocol for direct bacterial identification from positive blood cultures by using matrix assisted laser desorption ionization time-of- flight mass spectrometry. BMC Microbiol 15:258
Fitzgerald C, Stapleton P, Phelan E, Mulhare P, Carey B, Hickey M, Lynch B (2016 Apr) Doyle M (2016) rapid identification and antimicrobial susceptibility testing of positive blood cultures using MALDI-TOF MS and a modification of the standardised disc diffusion test: a pilot study. J Clin Pathol 27
Verroken A, Defourny L, le Polain de Waroux O, Belkhir L, Laterre PF, Delmee M, Glupczynski Y (2016) Clinical impact of MALDI-TOF MS identification and rapid susceptibility testing on adequate antimicrobial treatment in Sepsis with positive blood cultures. PLoS One 11(5):e0156299
Kim JM, Kim I, Chung SH, Chung Y, Han M, Kim JS (2019) Rapid discrimination of methicillin-resistant Staphylococcus aureus by MALDI-TOF MS. Pathogens 8(4)
Axelsson C, Rehnstam-Holm AS, Nilson B (2019) Rapid detection of antibiotic resistance in positive blood cultures by MALDI-TOF MS and an automated and optimized MBT-ASTRA protocol for Escherichia coli and Klebsiella pneumoniae. Infect Dis (Lond):1–9
Cherkaoui A, Cherpillod P, Renzi G, Schrenzel J, Kaiser L, Schibler M (2019) A molecular based diagnosis of positive blood culture in the context of viral haemorrhagic fever: proof of concept. Clin Microbiol Infect 25(10):1289 e1281–1289 e1284
Huang TD, Melnik E, Bogaerts P, Evrard S, Glupczynski Y (2019) Evaluation of the ePlex blood culture identification panels for detection of pathogens in bloodstream infections. J Clin Microbiol 57(2)
Kolb M, Lazarevic V, Emonet S, Calmy A, Girard M, Gaia N, Charretier Y, Cherkaoui A, Keller P, Huber C, Schrenzel J (2019) Next-generation sequencing for the diagnosis of challenging culture-negative endocarditis. Front Med (Lausanne) 6:203
Lazarevic V, Gaia N, Girard M, Leo S, Cherkaoui A, Renzi G, Emonet S, Jamme S, Ruppe E, Vijgen S, Rubbia-Brandt L, Toso C, Schrenzel J (2018) When bacterial culture fails, Metagenomics can help: a case of chronic hepatic Brucelloma assessed by next-generation sequencing. Front Microbiol 9:1566
Acknowledgment
The authors would like to thank Stéfane MARQUES MELANCIA, Marc JACQUES VINCENT, and Adrien FISCHER for technical assistance.
Funding
This study was supported using internal funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
JS received restricted research grants and participated to advisory boards from BioMérieux and Debiopharm. NV received restricted research grants from Roche. This research was conducted in the absence of any commercial or financial relationships in the last 3 years that could be construed as a potential conflict of interest.
Ethical approval
In accordance with the local ethical committee (Commission cantonale d’éthique de la recherche, https://www.hug-ge.ch/ethique), routine clinical laboratories of our institution may use biological sample leftovers for method development after irreversible anonymization of the data.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cherkaoui, A., Renzi, G., Azam, N. et al. Rapid identification by MALDI-TOF/MS and antimicrobial disk diffusion susceptibility testing for positive blood cultures after a short incubation on the WASPLab. Eur J Clin Microbiol Infect Dis 39, 1063–1070 (2020). https://doi.org/10.1007/s10096-020-03817-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-03817-8