Abstract
Significant advances have been made in the molecular assays used for the detection of human immunodeficiency virus (HIV), which are crucial in preventing HIV transmission and monitoring disease progression. Molecular assays for HIV diagnosis have now reached a high degree of specificity, sensitivity and reproducibility, and have less operator involvement to minimize risk of contamination. Furthermore, analyses have been developed for the characterization of host gene polymorphisms and host responses to better identify and monitor HIV-1 infections in the clinic. Currently, molecular technologies including HIV quantitative and qualitative assays are mainly based on the polymerase chain reaction (PCR), transcription-mediated amplification (TMA), nucleic acid sequence-based amplification (NASBA), and branched chain (b) DNA methods and widely used for HIV detection and characterization, such as blood screening, point-of-care testing (POCT), pediatric diagnosis, acute HIV infection (AHI), HIV drug resistance testing, antiretroviral (AR) susceptibility testing, host genome polymorphism testing, and host response analysis. This review summarizes the development and the potential utility of molecular assays used to detect and characterize HIV infections.
Similar content being viewed by others
References
(2018) HIV/AIDS Fact sheets, http://www.who.int/en/news-room/fact-sheets/detail/hiv-aids
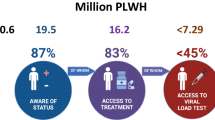
UNAIDS DATA 2017 http://www.unaids.org/en/resources/documents/2017/2017_data_book
Branson B, Owen SM, Bennett B, Werner BG, Pentella MA, Wesolowski LG (2014) Laboratory testing for the diagnosis of HIV infection: updated recommendations. Algorithms
Earl LA, Lifson JD, Subramaniam S (2013) Catching HIV ‘in the act’ with 3D electron microscopy. Trends Microbiol 21(8):397–404
Viana IFT, Coelho DF, Palma ML, Nascimento EJM, Gu G, Lima LFO et al (2018) Detection of IgG3 antibodies specific to the human immunodeficiency virus type 1 (HIV-1) p24 protein as marker for recently acquired infection. Epidemiol Infect 146(10):1293–1300
Ou CY, Kwok S, Mitchell SW, Mack DH, Sninsky JJ, Krebs JW et al (1988) DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science 239(4837):295–297
Brauer M, De Villiers JC, Mayaphi SH (2013) Evaluation of the determine fourth generation HIV rapid assay. J Virol Methods 189(1):180–183
Ly TD, Laperche S, Brennan C, Vallari A, Ebel A, Hunt J et al (2004) Evaluation of the sensitivity and specificity of six HIV combined p24 antigen and antibody assays. J Virol Methods 122(2):185–194
Zhao Y, Gou Y, Li D, Wang T, Huang X, Shi M et al (2018) Performance evaluation of a new automated fourth-generation HIV Ag/Ab combination chemiluminescence immunoassay. Clin Chem Lab Med 56(5):e115–e117
Delaney KP, Wesolowski LG, Owen SM (2017) The evolution of HIV testing continues. Sex Transm Dis 44(12):747–749
(2018) Advantages and disadvantages of FDA-approved HIV assays used for screening, by test category, https://www.cdc.gov/hiv/pdf/testing/hiv-tests-advantages-disadvantages_1.pdf
(2014) Laboratory testing for the diagnosis of hiv infection updated recommendations, https://stacks.cdc.gov/view/cdc/23447
Branson BM, Stekler JD (2012) Detection of acute HIV infection: we can’t close the window. J Infect Dis 205(4):521–524
Weber B (2006) Screening of HIV infection: role of molecular and immunological assays. Expert Rev Mol Diagn 6(3):399–411
Galel SA, Simon TL, Williamson PC, AuBuchon JP, Waxman DA, Erickson Y et al (2018) Sensitivity and specificity of a new automated system for the detection of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus nucleic acid in blood and plasma donations. Transfusion 58(3):649–659
Pas S, Rossen JW, Schoener D, Thamke D, Pettersson A, Babiel R et al (2010) Performance evaluation of the new Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test version 2.0 for quantification of human immunodeficiency virus type 1 RNA. J Clin Microbiol 48(4):1195–1200
Schmidt M, Korn K, Nubling CM, Chudy M, Kress J, Horst HA et al (2009) First transmission of human immunodeficiency virus type 1 by a cellular blood product after mandatory nucleic acid screening in Germany. Transfusion 49(9):1836–1844
Müller B, Nübling CM, Kress J, Roth WK, De Zolt S, Pichl L (2013) How safe is safe: new human immunodeficiency virus type 1 variants missed by nucleic acid testing. Transfusion 53(10pt2):2422–2430
Complete list of donor screening assays for infectious agents and HIV diagnostic assays, https://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm080466.htm#HIV1_NucleicAcid_Assays
Human immunodeficiency virus, type 1 (HIV-1) REVERSE TRANSCRIPTION (RT) polymerase chain reaction (PCR) assay, https://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm093501.htm
UltraQual HIV-1 RT-PCR Assay, https://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm172937.htm
COBAS AmpliScreen HIV-1 Test, https://www.fda.gov/downloads/BiologicsBloodVaccines/Blood-BloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM093531.pdf
APTIMA HIV-1 RNA Qualitative Assay, https://www.fda.gov/downloads/BiologicsBloodVaccines-/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM149927.pdf
Aptima HIV-1 Quant Assay, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/UCM534609.pdf
cobas HIV-1 Quantitative nucleic acid test for use on the cobas 6800/8800 Systems, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/UCM478438.pdf
Roche Amplicor HIV-1 Monitor Test, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/UCM093317.pdf
COBAS AmpliPrep/COBAS TaqMan HIV-1 Test, 48 Tests; COBAS AmpliPrep/COBAS TaqMan Wash Reagent, 5.1 L, https://www.fda.gov/downloads/ BiologicsBloodVaccines/BloodBloodProducts/Approv-edProducts/PremarketApprovalsPMAs/UCM092878.pdf
Abbott RealTime HIV-1 Amplification Reagent Kit, Abbott RealTime HIV-1 Calibrator Kit, Abbott RealTime HIV-1 Control Kit, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/UCM091193.pdf
Gomes P, Palma AC, Cabanas J, Abecasis A, Carvalho AP, Ziermann R et al (2006) Comparison of the COBAS TAQMAN HIV-1 HPS with VERSANT HIV-1 RNA 3.0 assay (bDNA) for plasma RNA quantitation in different HIV-1 subtypes. J Virol Methods 135(2):223–228
VERSANT HIV-1 RNA 3.0 Assay (bDNA), https://www.fda.gov/downloads/BiologicsBloodVaccines-/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/UCM091276.pdf
Eshleman SH, John H, Priscilla S, Cunningham SP, Birgit D, Catherine B et al (2004) Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System for sequence-based analysis of diverse human immunodeficiency virus type 1 strains. J Clin Microbiol 42(6):2711
ViroSeq HIV-1 Genotyping System, https://www.molecular.abbott/us/en/products/infectious-disease/viroseq-hiv-1-genotyping-system
Robert MG, Daniel RK, Victoria AJ, John WM, John LS, Ronald S et al (2003) Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol 41(4):1586
COBAS TaqScreen MPX Test, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloo-dProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM176443.pdf
cobas TaqScreen MPX Test, version 2.0 for use with the cobas s 201 system, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM427750.pdf
cobas MPX Test, for use on the cobas 6800/8800 Systems, https://www.fda.gov/downloads/Biol-ogicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM614913.pdf
Procleix Ultrio Assay, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProd-ucts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM335285.pdf
Procleix Ultrio Plus Assays, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBlood-Products/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM092120.pdf
Procleix Ultrio Elite Assay, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBlood-Products/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM606730.pdf
NGI UltraQual Multiplex PCR Assay for HCV, HIV-1, HIV-2 and HBV, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM609997.pdf
Bourlet T, Memmi M, Saoudin H, Pozzetto B (2013) Molecular HIV screening. Expert Rev Mol Diagn 13(7):693–705
Stevens WS, Noble L, Berrie L, Sarang S, Scott LE (2009) Ultra-high-throughput, automated nucleic acid detection of human immunodeficiency virus (HIV) for infant infection diagnosis using the Gen-Probe Aptima HIV-1 screening assay. J Clin Microbiol 47(8):2465–2469
Simonds RJ, Brown TM, Thea DM, Orloff SL, Steketee RW, Lee FK et al (1998) Sensitivity and specificity of a qualitative RNA detection assay to diagnose HIV infection in young infants. Perinatal AIDS Collaborative Transmission Study. Aids 12(12):1545–1549
Dubrow R, Qin L, Lin H, Hernandez-Ramirez RU, Neugebauer RS, Leyden W et al (2017) Association of CD4+ T-cell count, HIV-1 RNA viral load, and antiretroviral therapy with Kaposi sarcoma risk among HIV-infected persons in the United States and Canada. J Acquir Immune Defic Syndr 75(4):382–390
May MT, Gompels M, Delpech V, Porter K, Orkin C, Kegg S et al (2014) Impact on life expectancy of HIV-1 positive individuals of CD4+ cell count and viral load response to antiretroviral therapy. Aids 28(8):1193–1202
Mellors JW, Rinaldo CR Jr, Gupta P, White RM, Todd JA, Kingsley LA (1996) Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272(5265):1167–1170
Cozzi LA, Katzenstein TL, Ullum H, Phillips AN, Skinhøj P, Gerstoft J et al (1998) The relative prognostic value of plasma HIV RNA levels and CD4 lymphocyte counts in advanced HIV infection. Aids 12(13):1639–1643
Abbott MA, Poiesz BJ, Byrne BC, Kwok S, Sninsky JJ, Ehrlich GD (1988) Enzymatic gene amplification: qualitative and quantitative methods for detecting proviral DNA amplified in vitro. J Infect Dis 158(6):1158–1169
Malek L, Sooknanan R, Compton J (1994) Nucleic acid sequence-based amplification (NASBA). Methods Mol Biol 28(28):253
Horn T, Chang CA, Urdea MS (1997) Chemical synthesis and characterization of branched oligodeoxyribonucleotides (bDNA) for use as signal amplifiers in nucleic acid quantification assays. Nucleic Acids Res 25(23):4842–4849
Sloma CR, Germer JJ, Gerads TM, Mandrekar JN, Mitchell PS, Yao JD (2009) Comparison of the Abbott realtime human immunodeficiency virus type 1 (HIV-1) assay to the Cobas AmpliPrep/Cobas TaqMan HIV-1 test: workflow, reliability, and direct costs. J Clin Microbiol 47(4):889–895
Schumacher W, Frick E, Kauselmann M, Maier-Hoyle V, van der Vliet R, Babiel R (2007) Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol 38(4):304–312
Korn K, Weissbrich B, Henke-Gendo C, Heim A, Jauer CM, Taylor N et al (2009) Single-point mutations causing more than 100-fold underestimation of human immunodeficiency virus type 1 (HIV-1) load with the Cobas TaqMan HIV-1 real-time PCR assay. J Clin Microbiol 47(4):1238–1240
Tung YC, Ke LY, Lu PL, Lin KH, Lee SC, Lin YY et al (2015) Comparison of the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 test V1.0 with V2.0 in HIV-1 viral load quantification. Kaohsiung J Med Sci 31(4):188–193
Mourez T, Delaugerre C, Vray M, Lemee V, Simon F, Plantier JC (2015) Comparison of the bioMerieux NucliSENS EasyQ HIV-1 V2.0-HIV-1 RNA quantification assay versus Abbott RealTime HIV-1 and Roche Cobas TaqMan HIV-1 V2.0 on current epidemic HIV-1 variants. J Clin Virol 71:76–81
Manak MM, Hack HR, Nair SV, Worlock A, Malia JA, Peel SA et al (2016) Evaluation of Hologic Aptima HIV-1 Quant Dx assay on the Panther System on HIV subtypes. J Clin Microbiol 54(10):2575–2581
Loetscher P (2017) Performance evaluation of Cobas® HIV-1, a quantitative nucleic acid test for use on the Cobas® 6800/8800 systems. Journal of HIV and AIDS 3(1)
Wiesmann F, Ehret R, Naeth G, Daumer M, Fuhrmann J, Kaiser R et al (2018) Multicenter evaluation of two next-generation HIV-1 quantitation assays, Aptima Quant Dx and Cobas 6800, in comparison to the RealTime HIV-1 reference assay. J Clin Microbiol 56(10)
Elbeik T, Loftus RA, Beringer S (2014) Health care industries perspective of viral load assays: the VERSANT HIV-1 RNA 3.0 assay. Expert Rev Mol Diagn 2(3):275–285
Zhang L, Jin C, Jiang Z, Tang T, Jiang Y, Pan PL (2017) Comparison of commercial HIV-1 viral load tests by using proficiency test results in China, 2013–2015. Zhonghua Liu Xing Bing Xue Za Zhi 38(9):1231–1235
Baumeister MA, Zhang N, Beas H, Brooks JR, Canchola JA, Cosenza C et al (2012) A sensitive branched DNA HIV-1 signal amplification viral load assay with single day turnaround. PLoS One 7(3):e33295
Xu S, Song A, Nie J, Li X, Wang Y (2010) Performance of NucliSens HIV-1 EasyQ Version 2.0 compared with six commercially available quantitative nucleic acid assays for detection of HIV-1 in China. Mol Diagn Ther 14(5):305
(2016) BioMerieux SA alerts customers about potential inaccurate test results when using NucliSENS® easyMAG® magnetic silica for nucleic acid extraction, https://www.fda.gov/MedicalDevice-s/Safety/ListofRecalls/ucm516437.htm
Roth WK, Weber M, Seifried E (1999) Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet 353(9150):359
Susan AF, Takesha M, Julie AEN, William CM (2013) Validation of the gen-probe Aptima qualitative HIV-1 RNA assay for diagnosis of human immunodeficiency virus infection in infants. J Clin Microbiol 51(12):4137–4140
Lelie PN, van Drimmelen HA, Cuypers HT, Best SJ, Stramer SL, Hyland C et al (2010) Sensitivity of HCV RNA and HIV RNA blood screening assays. Transfusion 42(5):527–536
(2010) WHO Guidelines Approved by the Guidelines Review Committee. WHO Recommendations on the Diagnosis of HIV Infection in Infants and Children. World Health Organization World Health Organization., Geneva
Hamlyn E, Jones V, Porter K, Fidler S (2010) Antiretroviral treatment of primary HIV infection to reduce onward transmission. Curr Opin HIV AIDS 5(4):283–290
Dodd RY (2015) Transfusion-transmitted infections: testing strategies and residual risk. Isbt Science 9(1):1–5
Shyamala V (2015) Nucleic acid technology (NAT) testing for blood screening: impact of individual donation and Mini Pool-NAT testing on analytical sensitivity, screening sensitivity and clinical sensitivity. Isbt Science 9(2):315–324
Van Laethem K, Beuselinck K, Van Dooren S, De Clercq E, Desmyter J, Vandamme AM (1998) Diagnosis of human immunodeficiency virus infection by a polymerase chain reaction assay evaluated in patients harbouring strains of diverse geographical origin. J Virol Methods 70(2):153–166
Chudy M, Weber-Schehl M, Pichl L, Jork C, Kress J, Heiden M et al (2012) Blood screening nucleic acid amplification tests for human immunodeficiency virus type 1 may require two different amplification targets. Transfusion 52(2):431–439
Margaritis AR, Brown SM, Seed CR, Kiely P, D'Agostino B, Keller AJ (2007) Comparison of two automated nucleic acid testing systems for simultaneous detection of human immunodeficiency virus and hepatitis C virus RNA and hepatitis B virus DNA. Transfusion 47(10):1783–1793
Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C et al (2009) Comparison of the analytical and operational performance of two viral nucleic acid test blood screening systems: Procleix Tigris and cobas s 201. Transfusion 49(2):289–300
Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C et al (2009) Sensitivity of two hepatitis B virus, hepatitis C virus (HCV), and human immunodeficiency virus (HIV) nucleic acid test systems relative to hepatitis B surface antigen, anti-HCV, anti-HIV, and p24/anti-HIV combination assays in seroconversion panels. Transfusion 49(2):301–310
Grabarczyk P, van Drimmelen H, Kopacz A, Gdowska J, Liszewski G, Piotrowski D et al (2013) Head-to-head comparison of two transcription-mediated amplification assay versions for detection of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1 in blood donors. Transfusion 53(10 Pt 2):2512–2524
Grabarczyk P, Koppelman M, Boland F, Sauleda S, Fabra C, Cambie G et al (2015) Inclusion of human immunodeficiency virus type 2 (HIV-2) in a multiplex transcription-mediated amplification assay does not affect detection of HIV-1 and hepatitis B and C virus genotypes: a multicenter performance evaluation study. Transfusion 55(9):2246–2255
Heim A (2016) Evaluation of the Procleix Ultrio Elite Assay and the Panther-System for individual NAT screening of blood, hematopoietic stem cell, tissue and organ donors. Transfus Med Hemother 43(3):177–182
Le Corfec E, Le Pont F, Tuckwell HC, Rouzioux C, Costagliola D (1999) Direct HIV testing in blood donations: variation of the yield with detection threshold and pool size. Transfusion 39(10):1141–1144
Ha J, Park Y, Kim HS (2017) Evaluation of clinical sensitivity and specificity of hepatitis B virus (HBV), hepatitis C virus, and human immunodeficiency Virus-1 by cobas MPX: detection of occult HBV infection in an HBV-endemic area. J Clin Virol 96:60–63
Migueles SA, Connors M (2010) Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. Jama 304(2):194–201
Mendoza D, Johnson SA, Peterson BA, Natarajan V, Salgado M, Dewar RL et al (2012) Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood 119(20):4645–4655
Cock KMD, Fowler MG, Mercier E, Vincenzi ID, Saba J, Hoff E et al (2000) Prevention of mother-to-child HIV transmission in resource-poor countries: translating research into policy and practice. Jama 283(9):1175–1182
Luzuriaga K, Mofenson LM (2016) Challenges in the elimination of pediatric HIV-1 infection. N Engl J Med 374(8):761–770
Canals F, Masia M, Gutierrez F (2018) Developments in early diagnosis and therapy of HIV infection in newborns. Expert Opin Pharmacother 19(1):13–25
Stevens W, Sherman G, Downing R, Parsons LM, Ou CY, Crowley S et al (2008) Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: consensus statement on the performance of laboratory assays for early infant diagnosis. Open AIDS J 2:17–25
Ou C-Y, Fiscus S, Ellenberger D, Parekh B, Korhonen C, Nkengasong J et al (2012) Early diagnosis of HIV infection in the breastfed infant. Adv Exp Med Biol 743(743):51–65
Pierce VM, Neide B, Hodinka RL (2011) Evaluation of the Gen-Probe Aptima HIV-1 RNA qualitative assay as an alternative to Western blot analysis for confirmation of HIV infection. J Clin Microbiol 49(4):1642–1645
Alvarez P, Prieto L, Martin L, Obiang J, Avedillo P, Vargas A et al (2017) Evaluation of four commercial virological assays for early infant HIV-1 diagnosis using dried blood specimens. Pediatr Res 81(1–1):80–87
Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H et al (2016) Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. J Virol Methods 229:35–39
Hsiao NY, Dunning L, Kroon M, Myer L (2016) Laboratory evaluation of the Alere q point-of-care system for early infant HIV diagnosis. PLoS One 11(3):e0152672
Koopman JS, Jacquez JA, Welch GW, Simon CP, Foxman B, Pollock SM et al (1997) The role of early HIV infection in the spread of HIV through populations. J Acquir Immune Defic Syndr Hum Retrovirol 14(3):249–258
Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C et al (2007) High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis 195(7):951–959
Bassett IV, Chetty S, Giddy J, Reddy S, Bishop K, Lu Z et al (2015) Screening for acute HIV infection in South Africa: finding acute and chronic disease. HIV Med 12(1):46–53
Martin EG, Salaru G, Mohammed D, Coombs RW, Paul SM, Cadoff EM (2013) Finding those at risk: acute HIV infection in Newark, NJ. J Clin Virol 58:E24–E28
Emerson B, Plough K (2013) Detection of acute HIV-1 infections utilizing NAAT technology in Dallas, Texas. J Clin Virol 58(Suppl 1):e48–e53
Drancourt M, Michellepage A, Boyer S, Raoult D (2016) The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev 29(3):429
Engel N, Pant PN (2015) Qualitative research on point-of-care testing strategies and programs for HIV. Expert Rev Mol Diagn 15(1):71
Chang M, Steinmetzer K, Raugi DN, Smith RA, Ba S, Sall F et al (2017) Detection and differentiation of HIV-2 using the point-of-care Alere q HIV-1/2 detect nucleic acid test. J Clin Virol 97:22–25
Jani IV, Meggi B, Vubil A, Sitoe NE, Bhatt N, Tobaiwa O et al (2016) Evaluation of the whole-blood Alere Q NAT point-of-care RNA assay for HIV-1 viral load monitoring in a primary health care setting in Mozambique. J Clin Microbiol 54(8):2104–2108
Scott L, Gous N, Carmona S, Stevens W (2015) Laboratory evaluation of the Liat HIV Quant (IQuum) whole-blood and plasma HIV-1 viral load assays for point-of-care testing in South Africa. J Clin Microbiol 53(5):1616–1621
Murtagh M (2013) HIV / AIDS diagnostic technology landscape. 3rd edition
Pironti A, Walter H, Pfeifer N, Knops E, Lübke N, Büch J et al (2017) Determination of phenotypic resistance cutoffs from routine clinical data. J Acquir Immune Defic Syndr 74(5):e129–e137
Tsai HC, Chen IT, Lee SS, Chen YS (2018) HIV-1 genotypic drug resistance in patients with virological failure to single-tablet antiretroviral regimens in southern Taiwan. Infection & Drug Resistance 11:1061
Mayers DL, Baxter JD (2017) Clinical implications of HIV-1 drug resistance. In: Mayers DL, Sobel JD, Ouellette M, Kaye KS, Marchaim D (eds) Antimicrobial drug resistance: clinical and epidemiological aspects, Volume 2. Springer International Publishing, Cham, pp 1213–1225
Pattery T, Verlinden Y, De Wolf H, Nauwelaers D, Van Baelen K, Van Houtte M et al (2012) Development and performance of conventional HIV-1 phenotyping (Antivirogram(R)) and genotype-based calculated phenotyping assay (virco(R)TYPE HIV-1) on protease and reverse transcriptase genes to evaluate drug resistance. Intervirology 55(2):138–146
Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF et al (2013) Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382(9893):700–708
Westen GJPV, Hendriks A, Wegner JK, Ijzerman AP, Vlijmen HWTV, Bender A (2013) Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput Biol 9(2(2013-2-21)):e1002899
Mohamed S, Penaranda G, Gonzalez D, Camus C, Khiri H, Boulmé R et al (2014) Clinical impact of ultra deep versus Sanger sequencing detection of minority mutations on HIV-1 drug resistance genotype interpretation after virological failure. Aids 14(S2):1–1
Mekue MLC, Hélène P, Angélique NM, Donato K, Dieu LJD, François-Xavier MK et al (2015) LETTER TO THE EDITOR Performance of the ViroSeq® HIV-1 genotyping system V2.0 in Central Africa. Open Aids Journal 9(1):9–13
Paraschiv S, Otelea D, Baicus C, Tinischi M, Costache M, Neaga E (2009) Nucleoside reverse transcriptase inhibitor resistance mutations in subtype F1 strains isolated from heavily treated adolescents in Romania. Int J Infect Dis 13(1):81–89
Stelzl E, Proll J, Bizon B, Niklas N, Danzer M, Hackl C et al (2011) Human immunodeficiency virus type 1 drug resistance testing: evaluation of a new ultra-deep sequencing-based protocol and comparison with the TRUGENE HIV-1 genotyping kit. J Virol Methods 178(1–2):94–97
Duarte HA, Panpradist N, Beck IA, Lutz B, Lai J, Kanthula RM et al (2017) Current status of point-of-care testing for human immunodeficiency virus drug resistance. J Infect Dis 216(suppl_9):S824–s828
Metzner KJ, Rauch P, Braun P, Knechten H, Ehret R, Korn K et al (2011) Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol 50(2):156–161
Grant RM, Liegler T, Defechereux P, Kashuba AD, Taylor D, Abdelmohsen M et al (2015) Drug resistance and plasma viral RNA level after ineffective use of oral pre-exposure prophylaxis in women. Aids 29(3):331
Zhang G, Cai F, de Rivera IL, Zhou Z, Zhang J, Nkengasong J et al (2016) Simultaneous detection of major drug resistance mutations of HIV-1 subtype B viruses from dried blood spot specimens by multiplex allele-specific assay. J Clin Microbiol 54(1):220–222
Clutter DS, Rojas Sanchez P, Rhee SY, Shafer RW (2016) Genetic variability of HIV-1 for drug resistance assay development. Viruses 8(2)
Quinones-Mateu ME, Avila S, Reyes-Teran G, Martinez MA (2014) Deep sequencing: becoming a critical tool in clinical virology. J Clin Virol 61(1):9–19
Thiam M, Diop-Ndiaye H, Kebe K, Vidal N, Diakhate-Lô R, Diouara AA et al (2013) Performance of the ViroSeq HIV-1 genotyping system V2.0 on HIV-1 strains circulating in Senegal. J Virol Methods 188(1–2):97–103
Van Laethem K, Theys K, Vandamme AM (2015) HIV-1 genotypic drug resistance testing: digging deep, reaching wide? Curr Opin Virol 14:16–23
Johnson EO, Hancock DB, Gaddis NC, Levy JL, Page G, Novak SP et al (2015) Novel genetic locus implicated for HIV-1 acquisition with putative regulatory links to HIV replication and infectivity: a genome-wide association study. PLoS One 10(3):e0118149
McLaren PJ, Pulit SL, Gurdasani D, Bartha I, Shea PR, Pomilla C et al (2017) Evaluating the impact of functional genetic variation on HIV-1 control. J Infect Dis 216(9):1063–1069
Skerlj RT, Bridger GJ, Kaller A, Mceachern EJ, Crawford JB, Zhou Y et al (2010) Discovery of novel small molecule orally bioavailable C-X-C chemokine receptor 4 antagonists that are potent inhibitors of T-tropic (X4) HIV-1 replication. J Med Chem 53(8):3376–3388
Tsukamoto T (2018) Transcriptional gene silencing limits CXCR4-associated depletion of bone marrow CD34+ cells in HIV-1 infection. Aids 32(13):1737–1747
Liu Z, Chen S, Jin X, Wang Q, Yang K, Li C et al (2017) Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4 + T cells from HIV-1 infection. Cell Biosci 7(1):47
D'Antoni ML, Paul RH, Mitchell BI, Kohorn L, Fischer L, Lefebvre E et al (2018) Improved cognitive performance and reduced monocyte activation in virally suppressed chronic HIV following dual CCR2 and CCR5 antagonism. J Acquir Immune Defic Syndr 79(1):1
Ioannidis JPA, Rosenberg PS, Goedert JJ, Ashton LJ, Benfield TL, Buchbinder SP et al (2001) Effects of CCR5-Δ 32, CCR2-64I, and SDF-1 3′a alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med 135(9):782–795
Verheyen J, Thielen A, Lübke N, Dirks M, Widera M, Dittmer U, et al (2018) Rapid rebound of a preexisting CXCR4-tropic HIV variant after allogeneic transplantation with CCR5 delta32 homozygous stem cells. Clinical Infectious Diseases An Official Publication of the Infectious Diseases Society of America
Mori M, Wichukchinda N, Miyahara R, Rojanawiwat A, Pathipvanich P, Maekawa T et al (2014) HLA-B*35: 05 is a protective allele with a unique structure among HIV-1 CRF01_AE-infected Thais, in whom the B*57 frequency is low. Aids 28(7):959–967
Ragoussis J (2009) Genotyping technologies for genetic research. Annu Rev Genomics Hum Genet 10(1):117–133
Dumoulin A, Hirsch HH (2011) Reevaluating and optimizing polyomavirus BK and JC real-time PCR assays to detect rare sequence polymorphisms. J Clin Microbiol 49(4):1382
Komninakis S, Fukumori L, Alcalde R, Cortina M, Abdala L, Brito A et al (2007) Techniques used to identify the Brazilian variant of HIV-1 subtype B. Braz J Med Biol Res 40(3):301
Devadas K, Biswas S, Haleyurgirisetty M, Wood O, Ragupathy V, Lee S et al (2016) Analysis of host gene expression profile in HIV-1 and HIV-2 infected T-cells. PLoS One 11(1):e0147421
Lynch HE, Sempowski GD (2013) Molecular measurement of T cell receptor excision circles. Methods Mol Biol 979(979):147
Julia D, Nienke V, Tendai M, Bregje DBA, Otto SA, Hazenberg MD et al (2016) Reconciling longitudinal naive T-cell and TREC dynamics during HIV-1 infection. PLoS One 11(3):e0152513
Touloumi G, Pantazis N, Karafoulidou A, Mandalaki T, Goedert JJ, Kostrikis LG et al (2004) Changes in T cell receptor excision DNA circle (TREC) levels in HIV type 1-infected subjects pre- and post-highly active antiretroviral therapy. Aids Research & Human Retroviruses 20(1):47–54
Vogelstein B, Kinzler KW (1999) Digital PCR. Proc Natl Acad Sci U S A 96(16):9236–9241
Jones RB, Mueller S, O'Connor R, Rimpel K, Sloan DD, Karel D et al (2016) A subset of latency-reversing agents expose HIV-infected resting CD4+ T-cells to recognition by cytotoxic T-lymphocytes. PLoS Pathog 12(4):e1005545
Yucha RW, Hobbs KS, Hanhauser E, Hogan LE, Nieves W, Ozen MO et al (2017) High-throughput characterization of HIV-1 reservoir reactivation using a single-cell-in-droplet PCR assay. Ebiomedicine 20(C):217–229
Hancock G, Morónlópez S, Kopycinski J, Puertas MC, Giannoulatou E, Rose A et al (2017) Evaluation of the immunogenicity and impact on the latent HIV-1 reservoir of a conserved region vaccine, MVA.HIVconsv, in antiretroviral therapy-treated subjects. Journal of the International Aids Society 20(1):1
Mothe B, Climent N, Plana M, Rosà M, Luis J, Nez J et al (2015) Safety and immunogenicity of a modified vaccinia Ankara-based HIV-1 vaccine (MVA-B) in HIV-1-infected patients alone or in combination with a drug to reactivate latent HIV-1. J Antimicrob Chemother 70(6):1833–1842
Goo L, Jalalianlechak Z, Richardson BA, Overbaugh J (2012) A combination of broadly neutralizing HIV-1 monoclonal antibodies targeting distinct epitopes effectively neutralizes variants found in early infection. J Virol 86(19):10857–10861
Mccoy LE, Weiss RA (2013) Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 210(2):209–223
Stahl T, Böhme MU, Kröger N, Fehse B (2015) Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp Hematol 43(6):462–468.e461
Rosas-Umbert M, Mothe B, Noguera-Julian M, Bellido R, Puertas MC, Carrillo J et al (2017) Virological and immunological outcome of treatment interruption in HIV-1-infected subjects vaccinated with MVA-B. PLoS One 12(9):e0184929
Maria JB, Enrique M-G, Florencia P, Zhengyu O, Hong S, Jonathan ZL et al (2014) Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol 88(17):10056–10065
Chaillon A, Gianella S, Lada SM, Perez-Santiago J, Jordan P, Ignacio C et al (2017) Size, composition, and evolution of HIV DNA populations during early antiretroviral therapy and intensification with Maraviroc. J Virol 92(3):JVI.01589–JVI.01517
Bosman KJ, Nijhuis M, van Ham PM, Wensing AM, Vervisch K, Vandekerckhove L et al (2015) Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir. Sci Rep 5(2):13811
Rutsaert S, Bosman K, Trypsteen W, Nijhuis M, Vandekerckhove L (2018) Digital PCR as a tool to measure HIV persistence. Retrovirology 15(1):16
Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH et al (2013) Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8(4):e55943
Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR (2012) Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods 186(1–2):68–72
Ho Y, Shan L, Hosmane Nina N, Wang J, Laskey Sarah B, Rosenbloom Daniel IS et al (2013) Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155(3):540–551
Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M (1996) Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry 35(11):3362–3367
Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K et al (2009) Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360(7):692–698
Soriano V (2017) Hot news: gene therapy with CRISPR/Cas9 coming to age for HIV cure. AIDS Rev 19(3):167–172
Zulfiqar HF, Javed A, Sumbal, Afroze B, Ali Q, Akbar K et al (2017) HIV diagnosis and treatment through advanced technologies. Front Public Health 5:32
Zhu W, Lei R, Duff YL, Li J, Guo F, Wainberg MA et al (2015) The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 12(1):22
Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S et al (2016) CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep 15(3):481–489
Hütter G, Bodor J, Ledger S, Boyd M, Millington M, Tsie M et al (2015) CCR5 targeted cell therapy for HIV and prevention of viral escape. Viruses 7(8):4186–4203
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, J., Chang, L. & Wang, L. Nucleic acid testing and molecular characterization of HIV infections. Eur J Clin Microbiol Infect Dis 38, 829–842 (2019). https://doi.org/10.1007/s10096-019-03515-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03515-0