Abstract
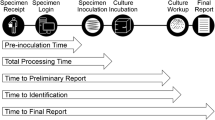
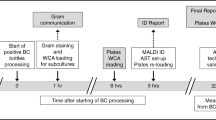
In order to realize the full potential of total laboratory automation (TLA) in the clinical microbiology laboratory, workflows must be optimized to match each laboratory’s capabilities, patient population, and staffing model. Using TLA-based digital photography to monitor urine cultures, we sought to improve culture result turn-around-time (TAT) by changing the time at which a culture is first photographed and thus available for analysis/work-up (Pre1) from 18 h (16,391 cultures) to 16 h (53,113 cultures) (with a total of 24-h culture incubation in both time periods); in both time periods, cultures were set up 24/7, and culture work-up occurred during the day shift only. With this change, we observed a significant decrease in time-to-final-result TAT for positive cultures (18 h-Pre1 median: 71.6 h; 16 h-Pre1 median: 61.0 h). This effect was most pronounced for Gram-negative organisms, with a median reduction in time-to-final-result for Escherichia coli cultures (51.8% of positive urine cultures) of 14.2 h (18 h-Pre1 median: 77.3 h; 16 h-Pre1 median: 63.1 h). This reduction in TAT was accompanied by a decrease in sensitivity at the Pre1 time point (18 h-Pre1 91.01%; 16 h-Pre1 88.06%), which we also found to vary by species: there was a reduction in sensitivity at the first culture reading of 1 to 2% for cultures with Gram-negative microorganisms, but for some Gram-positive microorganisms (e.g., Aerococcus urinae and non-aureus Staphylococcus species), there was a reduction in sensitivity at the Pre1 time-point of 3 to 7%. These results can guide workflow decisions for laboratories seeking to implement and/or optimize TLA systems, demonstrating a tradeoff between TAT and the sensitivity of preliminary urine culture results.




Similar content being viewed by others
References
Greub G, Prod’hom G (2011) Automation in clinical bacteriology: what system to choose. Clin Microbiol Infect 17:655–660
Bailey A, Burnham C-A, Ledeboer N (2018) Clinical microbiology is growing up: the Total Laboratory automation revolution. Clin 2017:274522
Dauwalder O, Landrieve L, Laurent F, de Montclos M, Vandenesch F, Lina G (2016) Does bacteriology laboratory automation reduce time to results and increase quality management. Clin Microbiol Infect 22:236–243
Mutters NT, Hodiamont CJ, de Jong MD, Overmeijer HP, van den Boogaard M, Visser CE (2014) Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann Lab Med 34:111–117
Yarbrough ML, Lainhart W, McMullen AR, Anderson NW, Burnham CD (2018) Impact of total laboratory automation on workflow and specimen processing time for culture of urine specimens. Eur J Clin Microbiol Infect Dis 37(12):2405–2411
Theparee T, Das S, Thomson RB (2018) Total Laboratory automation and matrix-assisted laser desorption ionization-time of flight mass spectrometry improve turnaround times in the clinical microbiology laboratory: a retrospective analysis. J Clin Microbiol 26:56(1)
Burckhardt I, Last K, Zimmermann S (2019) Shorter incubation times for detecting multi-drug resistant Bacteria in patient samples: defining early imaging time points using growth kinetics and Total Laboratory automation. Ann Lab Med 39:43–49
Foxman B (2014) Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin N Am 28:1–13
Lainhart W, Burnham CA (2018) Enhanced recovery of fastidious organisms from urine culture in the setting of Total Laboratory automation. J Clin Microbiol 26:56(6). https://doi.org/10.1128/JCM.00546-18
Klein S, Nurjadi D, Horner S, Heeg K, Zimmermann S, Burckhardt I (2018) Significant increase in cultivation of Gardnerella vaginalis, Alloscardovia omnicolens, Actinotignum schaalii, and Actinomyces spp. in urine samples with total laboratory automation. Eur J Clin Microbiol Infect Dis 37:1305–1311
De Socio GV, Di Donato F, Paggi R, Gabrielli C, Belati A, Rizza G, Savoia M, Repetto A, Cenci E, Mencacci A (2018) Laboratory automation reduces time to report of positive blood cultures and improves management of patients with bloodstream infection. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-018-3377-5
Burckhardt I, Horner S, Burckhardt F, Zimmermann S (2018) Detection of MRSA in nasal swabs-marked reduction of time to report for negative reports by substituting classical manual workflow with total lab automation. Eur J Clin Microbiol Infect Dis 37:1745–1751
Graham M, Tilson L, Streitberg R, Hamblin J, Korman TM (2016) Improved standardization and potential for shortened time to results with BD Kiestra™ total laboratory automation of early urine cultures: a prospective comparison with manual processing. Diagn Microbiol Infect Dis 86:1–4
Acknowledgements
CAB has received honoraria from BD and ASM for development of educational lectures related to TLA. The other authors declare that they have no conflicts of interest. The authors would like to thank the dedicated staff of the clinical microbiology laboratory at Barnes-Jewish Hospital for their ongoing work for our patients, and Abby Crozier for her efforts in Kiestra implementation and for critical reading of the manuscript. The authors would also like to thank the laboratory information systems team at Barnes-Jewish Hospital, in particular Barbara Banks, for help with procuring data from Cerner Millennium. ALB performed this work with support from the Physician Scientist Training Program (PSTP) sponsored by the Department of Pathology and Immunology at Washington University in St. Louis School of Medicine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics statement
This study was reviewed and approved by the Institutional Review Board at Washington University in St. Louis (IRB ID#: 201801212).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(XLSX 44 kb)
Rights and permissions
About this article
Cite this article
Bailey, A.L., Burnham, CA.D. Reducing the time between inoculation and first-read of urine cultures using total lab automation significantly reduces turn-around-time of positive culture results with minimal loss of first-read sensitivity. Eur J Clin Microbiol Infect Dis 38, 1135–1141 (2019). https://doi.org/10.1007/s10096-019-03512-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03512-3