Abstract
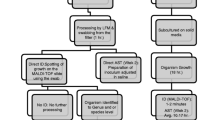
To standardize the methodology for conducting direct antimicrobial susceptibility testing (AST) of Enterobacterales and Pseudomonas aeruginosa causing bacteremia from positive blood culture pellets. Two methods for processing positive blood cultures with Enterobacterales and P. aeruginosa were compared: a conventional method for identification and AST versus a direct method obtaining a pellet for both matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) identification and direct AST. A total of 157 (145 Enterobacterales, 12 P. aeruginosa) positive blood cultures were included. Microorganism identification showed 100% concordance between both methods at species and genus level. Definitive AST results were obtained 24 h earlier with the rapid method than the conventional one (p < 0.001). Of the 2814 MICs generated, there were discrepancies with respect to the conventional method in 47 (1.7%), 0.3% being very major (VME) and 1.3% major (ME) errors. Better results for AST were obtained when colony counts with the pellet were ≥ 105 cfu/ml. The essential agreement (EA) for antibiotics tested in Enterobacterales was at least 97%, except for ampicillin (95%). Regardless of colony count, the greatest discrepancies were observed for first/s-generation cephalosporins and aminoglycosides. In P. aeruginosa, EA was at least 92%, except for piperacillin-tazobactam (84%) and cefepime (76%). No VME occurred except for ceftazidime (8%). ME occurred in piperacillin/tazobactam (16%), ticarcillin, ceftazidime, tobramycin, amikacin, and colistin (8% each). Direct use of the blood culture pellet permits fast AST in bacteremia of Enterobacterales, enabling the clinicians to perform an early treatment adjustment. However, for Pseudomonas aeruginosa, the data needs expanding to improve the reliability of this technique.


Similar content being viewed by others
References
Goff DA, Jankowski C, Tenover FC (2012) Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy 32:677–687. https://doi.org/10.1002/j.1875-9114.2012.01137.x
Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH (2000) The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146–155. https://doi.org/10.1378/chest.118.1.146
Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA et al (2007) Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother 19(61):428–435. https://doi.org/10.1093/jac/dkm497
2018,Antimicrobial stewardship: from principles to practice. BSAC
Rodríguez-Baño J, Cisneros JM, Cobos-Trigueros N, Fresco G, Navarro-San Francisco C, Gudiol C et al (2015) Diagnosis and antimicrobial treatment of invasive infections due to multidrug-resistant Enterobacteriaceae. Guidelines of the Spanish Society of Infectious Diseases and Clinical Microbiology. Enferm Infecc Microbiol Clin 33:337.e1–337.e21. https://doi.org/10.1016/j.eimc.2014.11.009
van den Bijllaardt W, Buiting AG, Mouton JW, Muller AE (2017) Shortening the incubation time for antimicrobial susceptibility testing by disk diffusion for Enterobacteriaceae: how short can it be and are the results accurate? Int J Antimicrob Agents 49:631–637. https://doi.org/10.1016/j.ijantimicag.2016.12.019
Chandrasekaran S, Abbott A, Campeau S, Zimmer BL, Weinstein M, Thrupp L et al (2018) Direct-from-blood-culture disk diffusion to determine antimicrobial susceptibility of gram-negative bacteria: preliminary report from the clinical and laboratory standards institute methods development and standardization working group. J Clin Microbiol 56:1071. https://doi.org/10.1128/JCM.01678-17
Descours G, Desmurs L, Hoang TLT, Ibranosyan M, Baume M, Ranc A-G et al (2018) Evaluation of the Accelerate Pheno™ system for rapid identification and antimicrobial susceptibility testing of Gram-negative bacteria in bloodstream infections. Eur J Clin Microbiol Infect Dis 28(15):395–311. https://doi.org/10.1007/s10096-018-3287-6
Korber F, Zeller I, Grünstäudl M, Willinger B, Apfalter P, Hirschl AM et al (2017) SeptiFast versus blood culture in clinical routine – a report on 3 years experience. Wien Klin Wochenschr 129:427–434. https://doi.org/10.1007/s00508-017-1181-3
Huang T-H, Tzeng Y-L, Dickson RM (2018) FAST: rapid determinations of antibiotic susceptibility phenotypes using label-free cytometry. Cytometry Part A 7(93):639–648. https://doi.org/10.1002/cyto.a.23370
Costa-de-Oliveira S, Teixeira-Santos R, Silva AP, Pinho E, Mergulhão P, Silva-Dias A et al (2017) Potential impact of flow cytometry antimicrobial susceptibility testing on the clinical management of gram-negative bacteremia using the FASTinov® Kit. Front Microbiol 12(8):167. https://doi.org/10.3389/fmicb.2017.02455
Prod'hom G, Durussel C, Greub G (2013) A simple blood-culture bacterial pellet preparation for faster accurate direct bacterial identification and antibiotic susceptibility testing with the VITEK 2 system. J Med Microbiol 19(62):773–777. https://doi.org/10.1099/jmm.0.049361-0
Clark RB, Lewinski MA, Loeffelholz MJ, Tibbetts RJ (2009) Cumitech 31A, verification and validation of procedures in the clinical microbiology laboratory. ASM Press, Washington, DC
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159. https://doi.org/10.2307/2529310
Ruiz-Aragón J, Ballestero-Téllez M, Gutiérrez-Gutiérrez B, De Cueto M, Rodríguez-Baño J, Pascual A (2018) Direct bacterial identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry: a systematic review and meta-analysis. Enferm Infecc Microbiol Clin 36:484–492. https://doi.org/10.1016/j.eimc.2017.08.012
Rosa R, Zavala B, Cain N, Anjan S, Aragon L, Abbo LM (2018) Antimicrobial stewardship program implementation of a quality improvement intervention using real-time feedback and an electronic order set for the management of Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol 39:346–349. https://doi.org/10.1017/ice.2017.325
Pogue JM, Mynatt RP, Marchaim D, Zhao JJ, Barr VO, Moshos J et al (2014) Automated alerts coupled with antimicrobial stewardship intervention lead to decreases in length of stay in patients with gram-negative bacteremia. Infect Control Hosp Epidemiol 35:132–138. https://doi.org/10.1086/674849
Bhowmick T, Kirn TJ, Hetherington F, Takavarasha S, Sandhu SS, Gandhi S et al (2018) Collaboration between an antimicrobial stewardship team and the microbiology laboratory can shorten time to directed antibiotic therapy for methicillin-susceptible staphylococcal bacteremia and to discontinuation of antibiotics for coagulase-negative staphylococcal contaminants. Diagn Microbiol Infect Dis:29. https://doi.org/10.1016/j.diagmicrobio.2018.05.020
Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J et al (2013) Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 57:1237–1245. https://doi.org/10.1093/cid/cit498
Köck R, Wüllenweber J, Horn D, Lanckohr C, Becker K, Idelevich EA (2017) Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control 14(6):353. https://doi.org/10.1186/s13756-017-0173-4
Richter SS, Ferraro MJ. 2011 Susceptibility testing instrumentation and computerized expert systems for data analysis and interpretation. In: Manual of clinical microbiology, 10th Edition. American Society of Microbiology, p. 1144–54.doi: https://doi.org/10.1128/9781555816728.ch69
Waites KB, Brookings ES, Moser SA, Zimmer BL (1998) Direct bacterial identification from positive BacT/alert blood cultures using MicroScan overnight and rapid panels. Diagn Microbiol Infect Dis 32:21–26. https://doi.org/10.1016/S0732-8893(98)00058-3
Fontanals D, Salceda F, Hernández J, Sanfeliu I, Torra M (2002) Evaluation of wider system for direct identification and antimicrobial susceptibility testing of gram-negative bacilli from positive blood culture bottles. Eur J Clin Microbiol Infect Dis 21:693–695. https://doi.org/10.1007/s10096-002-0791-4
De Cueto M, Ceballos E, Martínez-Martínez L, Perea EJ, Pascual A (2004) Use of positive blood cultures for direct identification and susceptibility testing with the vitek 2 system. J Clin Microbiol 42:3734–3738. https://doi.org/10.1128/JCM.42.8.3734-3738.2004
Parkins MD, Gregson DB, Pitout JDD, Ross T, Laupland KB (2009) Population-based study of the epidemiology and the risk factors for pseudomonas aeruginosa bloodstream infection. Infection 12(38):25–32. https://doi.org/10.1007/s15010-009-9145-9
Bruins MJ, Bloembergen P, Ruijs GJHM, Wolfhagen MJHM (2004) Identification and susceptibility testing of enterobacteriaceae and pseudomonas aeruginosa by direct inoculation from positive BACTEC blood culture bottles into Vitek 2. J Clin Microbiol 8(42):7–11. https://doi.org/10.1128/JCM.42.1.7-11.2004
Acknowledgements
RC research is partially supported by Plan Nacional de I+D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0011)—co-financed by European Development Regional Fund “A way to achieve Europe”, Operative program Intelligent Growth 2014–2020, and Fundación Soria Melguizo (Madrid, Spain).
We thank Mary Harper for English correction of the manuscript. Also, we thank Francisca Pérez, Maria Isabel Moya, and Isabel Soler for their daily work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the local ethics committee as it stated in the record number 332 of this committee.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
López-Pintor, J.M., Navarro-San Francisco, C., Sánchez-López, J. et al. Direct antimicrobial susceptibility testing from the blood culture pellet obtained for MALDI-TOF identification of Enterobacterales and Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 38, 1095–1104 (2019). https://doi.org/10.1007/s10096-019-03498-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03498-y