Abstract
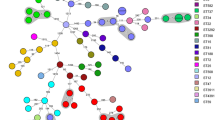
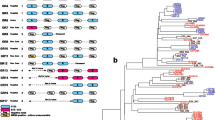
We describe a nosocomial outbreak of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) ST59-SCCmec type V in a neonatal intensive care unit (NICU) in Hong Kong. In-depth epidemiological analysis was performed by whole-genome sequencing (WGS) of the CA-MRSA isolates collected from patients and environment during weekly surveillance and healthcare workers from the later phase of the outbreak. Case–control analysis was performed to analyze potential risk factors for the outbreak. The outbreak occurred from September 2017 to February 2018 involving 15 neonates and one healthcare worker. WGS analysis revealed complicated transmission dynamics between patients, healthcare worker, and environment, from an unrecognized source introduced into the NICU within 6 months before the outbreak. In addition to enforcement of directly observed hand hygiene, environmental disinfection, cohort nursing of colonized and infected patients, together with contact tracing for secondary patients, medical, nursing, and supporting staff were segregated where one team would care for CA-MRSA-confirmed/CA-MRSA-exposed patients and the other for newly admitted patients in the NICU only. Case–control analysis revealed use of cephalosporins [odds ratio 49.84 (3.10–801.46), p = 0.006] and length of hospitalization [odds ratio 1.02 (1.00–1.04), p = 0.013] as significant risk factors for nosocomial acquisition of CA-MRSA in NICU using multivariate analysis. WGS facilitates the understanding of transmission dynamics of an outbreak, providing insights for outbreak prevention.




Similar content being viewed by others
References
Otter JA, French GL (2011) Community-associated meticillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated infection. J Hosp Infect 79(3):189–193
Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesoe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. (2018) SCCmecFinder, a Web-Based Tool for Typing of Staphylococcal CassetteChromosome mec in Staphylococcus aureus Using Whole-Genome Sequence Data. mSphere, 3(1): e00612-17
Sassi M, Felden B, Revest M, Tattevin P, Augagneur Y, Donnio PY (2017) An outbreak in intravenous drug users due to USA300 Latin-American variant community-acquired methicillin-resistant Staphylococcus aureus in France as early as 2007. Eur J Clin Microbiol Infect Dis 36(12):2495–2501
Cheng VC, Lau YK, Lee KL, Yiu KH, Chan KH, Ho PL, Yuen KY (2009) Fatal co-infection with swine origin influenza virus a/H1N1 and community-acquired methicillin-resistant Staphylococcus aureus. J Inf Secur 59(5):366–370
Ho PL, Cheung C, Mak GC, Tse CW, Ng TK, Cheung CH, Que TL, Lam R, Lai RW, Yung RW, Yuen KY (2007) Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn Microbiol Infect Dis 57(2):145–151
Ho PL, Chuang SK, Choi YF, Lee RA, Lit AC, Ng TK, Que TL, Shek KC, Tong HK, Tse CW, Tung WK, Yung RW, CAMsn HK (2008) Community-associated methicillin-resistant and methicillin-sensitive Staphylococcus aureus: skin and soft tissue infections in Hong Kong. Diagn Microbiol Infect Dis 61(3):245–250
Ho PL, Lo PY, Chow KH, Lau EH, Lai EL, Cheng VC, Kao RY (2010) Vancomycin MIC creep in MRSA isolates from 1997 to 2008 in a healthcare region in Hong Kong. J Inf Secur 60(2):140–145
Ho PL, Chiu SS, Chan MY, Gan Y, Chow KH, Lai EL, Lau YL (2012) Molecular epidemiology and nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus among young children attending day care centers and kindergartens in Hong Kong. J Inf Secur 64(5):500–506
Cheng VC, Tai JW, Wong LM, Ching RH, Ng MM, Ho SK, Lee DW, Li WS, Lee WM, Sridhar S, Wong SC, Ho PL, Yuen KY (2015) Effect of proactive infection control measures on benchmarked rate of hospital outbreaks: an analysis of public hospitals in Hong Kong over 5 years. Am J Infect Control 43(9):965–970
Cheng VCC, Wong SC, Chen JHK, So SYC, Wong SCY, Ho PL, Yuen KY (2018) Control of multidrug-resistant Acinetobacter baumannii in Hong Kong: role of environmental surveillance in communal areas after a hospital outbreak. Am J Infect Control 46(1):60–66
Cheng VC, Tai JW, Ng ML, Chan JF, Wong SC, Li IW, Chung HP, Lo WK, Yuen KY, Ho PL (2012) Extensive contact tracing and screening to control the spread of vancomycin-resistant Enterococcus faecium ST414 in Hong Kong. Chin Med J 125(19):3450–3457
Cheng VCC, Chen JHK, Wong SCY, Leung SSM, So SYC, Lung DC, Lee WM, Trendell-Smith NJ, Chan WM, Ng D, To L, Lie AKW, Yuen KY (2016) Hospital outbreak of pulmonary and cutaneous Zygomycosis due to contaminated linen items from substandard laundry. Clin Infect Dis 62(6):714–721
Cheng VC, Li IW, Wu AK, Tang BS, Ng KH, To KK, Tse H, Que TL, Ho PL, Yuen KY (2008) Effect of antibiotics on the bacterial load of meticillin-resistant Staphylococcus aureus colonisation in anterior nares. J Hosp Infect 70(1):27–34
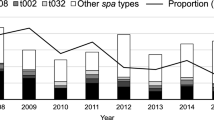
Cheng VC, Chan JF, Lau EH, Yam WC, Ho SK, Yau MC, Tse EY, Wong AC, Tai JW, Fan ST, Ho PL, Yuen KY (2011) Studying the transmission dynamics of meticillin-resistant Staphylococcus aureus in Hong Kong using spa typing. J Hosp Infect 79(3):206–210
Cheng VC, Tai JW, Wong ZS, Chen JH, Pan KB, Hai Y, Ng WC, Chow DM, Yau MC, Chan JF, Wong SC, Tse H, Chan SS, Tsui KL, Chan FH, Ho PL, Yuen KY (2013) Transmission of methicillin-resistant Staphylococcus aureus in the long term care facilities in Hong Kong. BMC Infect Dis 13:205
Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, Otto TD, Keane JA (2016) Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom 2(8):e000083
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402
Ho PL, Wang Y, Liu MC, Lai EL, Law PY, Cao H, Chow KH. (2018) IncX3 Epidemic Plasmid Carrying blaNDM-5 in Escherichia coli from Swine in Multiple Geographic Areas in China. Antimicrob Agents Chemother, 62(3): e02295-17
Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM (2014) Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52(5):1501–1510
Kossow A, Kampmeier S, Schaumburg F, Knaack D, Moellers M, Mellmann A. (2018) Whole genome sequencing reveals a prolonged and spatially spread nosocomial outbreak of Panton-Valentine leucocidinpositive meticillin-resistant Staphylococcus aureus (USA300). J Hosp Infect S0195-6701(18):30486-9
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573
Treangen TJ, Ondov BD, Koren S, Phillippy AM (2014) The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15(11):524
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4(1):vey016
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Didelot X, Fraser C, Gardy J, Colijn C (2017) Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol Biol Evol 34(4):997–1007
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian Phylogenetics using tracer 1.7. Syst Biol 67(5):901–904
Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14(7):1394–1403
Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A (2018) MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14(1):e1005944
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR (2015) Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43(3):e15
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274
Wang Y, Tong MK, Chow KH, Cheng VC, Tse CW, Wu AK, Lai RW, Luk WK, Tsang DN, Ho PL (2018) Occurrence of highly conjugative IncX3 epidemic plasmid carrying Bla NDM in Enterobacteriaceae isolates in geographically widespread areas. Front Microbiol 9:2272
Cheng VC, Wu AK, Cheung CH, Lau SK, Woo PC, Chan KH, Li KS, Ip IK, Dunn EL, Lee RA, Yam LY, Yuen KY (2007) Outbreak of human metapneumovirus infection in psychiatric inpatients: implications for directly observed use of alcohol hand rub in prevention of nosocomial outbreaks. J Hosp Infect 67(4):336–343
Cheng VCC, Wong SC, Wong SCY, Yuen KY. (2018) Directly observed hand hygiene - from healthcare workers to patients. J Hosp Infect S0195-6701(18):30671-6
Huang TW, Chen FJ, Miu WC, Liao TL, Lin AC, Huang IW, Wu KM, Tsai SF, Chen YT, Lauderdale TL (2012) Complete genome sequence of Staphylococcus aureus M013, a pvl-positive, ST59-SCCmec type V strain isolated in Taiwan. J Bacteriol 194(5):1256–1257
Cheng VC, Chen JH, Poon RW, Lee WM, So SY, Wong SC, Chau PH, Yip CC, Wong SS, Chan JF, Hung IF, Ho PL, Yuen KY (2015) Control of hospital endemicity of multiple-drug-resistant Acinetobacter baumannii ST457 with directly observed hand hygiene. Eur J Clin Microbiol Infect Dis 34(4):713–718
Cheng VC, Tai JW, Li WS, Chau PH, So SY, Wong LM, Ching RH, Ng MM, Ho SK, Lee DW, Lee WM, Wong SC, Yuen KY (2016) Implementation of directly observed patient hand hygiene for hospitalized patients by hand hygiene ambassadors in Hong Kong. Am J Infect Control 44(6):621–624
Cheng VC, Tai JW, Chau PH, Lai CK, Chuang VW, So SY, Wong SC, Chen JH, Ho PL, Tsang DN, Yuen KY (2016) Successful control of emerging vancomycin-resistant enterococci by territory-wide implementation of directly observed hand hygiene in patients in Hong Kong. Am J Infect Control 44(10):1168–1171
Cheng VC, To KK, Li IW, Tang BS, Chan JF, Kwan S, Mak R, Tai J, Ching P, Ho PL, Seto WH (2009) Antimicrobial stewardship program directed at broad-spectrum intravenous antibiotics prescription in a tertiary hospital. Eur J Clin Microbiol Infect Dis 28(12):1447–1456
Jernigan JA, Titus MG, Groschel DH, Getchell-White S, Farr BM (1996) Effectiveness of contact isolation during a hospital outbreak of methicillin-resistant Staphylococcus aureus. Am J Epidemiol 143(5):496–504
Sax H, Posfay-Barbe K, Harbarth S, Francois P, Touveneau S, Pessoa-Silva CL, Schrenzel J, Dharan S, Gervaix A, Pittet D (2006) Control of a cluster of community-associated, methicillin-resistant Staphylococcus aureus in neonatology. J Hosp Infect 63(1):93–100
Giuffre M, Cipolla D, Bonura C, Geraci DM, Aleo A, Di Noto S, Nociforo F, Corsello G, Mammina C (2012) Epidemic spread of ST1-MRSA-IVa in a neonatal intensive care unit, Italy. BMC Pediatr 12:64
Hawkins G, Stewart S, Blatchford O, Reilly J (2011) Should healthcare workers be screened routinely for meticillin-resistant Staphylococcus aureus? A review of the evidence. J Hosp Infect 77(4):285–289
Orendi JM, Coetzee N, Ellington MJ, Boakes E, Cookson BD, Hardy KJ, Hawkey PM, Kearns AM (2010) Community and nosocomial transmission of Panton-valentine leucocidin-positive community-associated meticillin-resistant Staphylococcus aureus: implications for healthcare. J Hosp Infect 75(4):258–264
Pierce R, Lessler J, Popoola VO, Milstone AM (2017) Meticillin-resistant Staphylococcus aureus (MRSA) acquisition risk in an endemic neonatal intensive care unit with an active surveillance culture and decolonization programme. J Hosp Infect 95(1):91–97
Feng Y, Chen HL, Chen CJ, Chen CL, Chiu CH (2017) Genome comparisons of two Taiwanese community-associated methicillin-resistant Staphylococcus aureus ST59 clones support the multi-origin theory of CA-MRSA. Infect Genet Evol 54:60–65
Hsu LY, Harris SR, Chlebowicz MA, Lindsay JA, Koh TH, Krishnan P, Tan TY, Hon PY, Grubb WB, Bentley SD, Parkhill J, Peacock SJ, Holden MT (2015) Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol 16:81
Fu Z, Ma Y, Chen C, Guo Y, Hu F, Liu Y, Xu X, Wang M (2015) Prevalence of Fosfomycin resistance and mutations in murA, glpT, and uhpT in methicillin-resistant Staphylococcus aureus strains isolated from blood and cerebrospinal fluid samples. Front Microbiol 6:1544
Acknowledgements
Genomic sequences of the CA-MRSA isolates in this work have been deposited in the GenBank under BioProject number PRJNA493547.
Funding
This study was partially supported by grants from the Health and Medical Research Fund (HMRF), Food and Health Bureau, Hong Kong SAR Government (HKM-15-M10 and HKM-15-M12), and Seed Fund for Basic Research, University Research Committee, University of Hong Kong.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethical approval was not required because the infection control measures are the standard patient care to control and prevent hospital outbreak.
Informed consent
No informed consent was required since this was a retrospective analysis of data.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cheng, V.C.C., Wong, SC., Cao, H. et al. Whole-genome sequencing data-based modeling for the investigation of an outbreak of community-associated methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in Hong Kong. Eur J Clin Microbiol Infect Dis 38, 563–573 (2019). https://doi.org/10.1007/s10096-018-03458-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-03458-y