Abstract
The aims of this study were to evaluate the frequency of Achromobacter xylosoxidans infection in a cohort of cystic fibrosis patients, to investigate antimicrobial sensitivity, to establish possible clonal likeness among strains, and to address the clinical impact of this infection or colonization on the general outcome of these patients. The study was undertaken between January 2004 and December 2008 on 300 patients receiving care at the Regional Cystic Fibrosis Center of the Naples University “Federico II”. Sputum samples were checked for bacterial identification. For DNA fingerprinting, pulsed-field gel electrophoresis (PFGE) was carried out. Fifty-three patients (17.6%) had at least one positive culture for A. xylosoxidans; of these, 6/53 (11.3%) patients were defined as chronically infected and all were co-colonized by Pseudomonas aeruginosa. Of the patients, 18.8% persistently carried multidrug-resistant isolates. Macrorestriction analysis showed the presence of seven major clusters. DNA fingerprinting also showed a genetic relationship among strains isolated from the same patients at different times. The results of DNA fingerprinting indicate evidence of bacterial clonal likeness among the enrolled infected patients. We found no significant differences in the forced expiratory volume in 1 s (FEV1) and body mass index (BMI) when comparing the case group of A. xylosoxidans chronically infected patients with the control group of P. aeruginosa chronically infected patients.
Similar content being viewed by others
Introduction
Over the past 20 years, the epidemiology of bacteria involved in acute infections in cystic fibrosis (CF) has become increasingly complex. Although Pseudomonas aeruginosa, Staphylococcus aureus, and Haemophilus influenzae have been the most common pathogens in the lower airways of CF patients, with improved survival, new emergent pathogens such as Burkholderia cepacia complex (Bcc), Stenotrophomonas maltophilia, Achromobacter xylosoxidans, Aspergillus spp., non-tuberculous mycobacteria, and respiratory viruses have been detected in the last few years [1–12]. Other unusual bacteria such as Acinetobacter spp., Bordetella spp., Moraxella spp., Comamonas spp., Rhizobium spp., Herbaspirillum spp., and Inquilinus limosus have recently been described [13], and, also, Italian studies have reported Gram-negative non-fermentative bacteria such as Chryseobacterium meningosepticum, Chryseobacterium indologenes, Chryseobacterium gleum, Sphingobacterium spiritivorum, and Sphingobacterium multivorum in sputum samples of CF patients [14, 15]. Moreover, mycetes such as Scedosporium apiospermum, Penicillium, and Exophiala have also been recognized [16–19]. The pathogenic role of several of these microorganisms has not yet been clarified.
A. xylosoxidans is an aerobic, oxidase- and catalase-positive, non-lactose fermenting, Gram-negative bacillus widely distributed in the natural environment. Its taxonomic position has been considered to be uncertain during the last decades: the genus was named Achromobacter, then Alcaligenes, and then again Achromobacter. The phylogenetic analyses of 16S rRNA, besides a difference of more than 10% in the GC content of DNA, demonstrated that A. xylosoxidans and Alcaligenes faecalis (type species of the genus Alcaligenes) belong to two distinct genera, Achromobacter and Alcaligenes, respectively [20].
A. xylosoxidans is an opportunistic human pathogen capable of causing a variety of infections, including bacteremia, meningitis, pneumonia, and peritonitis, particularly in immunocompromised hosts and patients with underlying diseases [21]. Nosocomial outbreaks attributed to disinfectant solutions, saline solutions, and diagnostic tracers contaminated with this species have been described [22, 23].
In patients with CF, an increasing prevalence of A. xylosoxidans isolates has recently been reported [24–28], but the clinical significance of A. xylosoxidans infection is still unclear.
Because of the relevance of bacterial lung infections in CF patients, the aims of our study were to: (1) evaluate the frequency of A. xylosoxidans infections in a cohort of CF patients, (2) investigate the antimicrobial sensitivity of isolates, (3) establish possible clonal likeness among strains, as well as to (4) elucidate possible clonal likeness among strains isolated from the same patients during the study period and (5) address a possible clinical relevance of this infection on the general outcome of these patients.
Patients and methods
Study population
A retrospective chart review of microbiological samples obtained from 300 CF patients (145 males and 155 females; mean age 16.21 years; range 0.5–50 years) regularly attending the Regional Referral Cystic Fibrosis Center of Naples, Italy, was undertaken. Sputum samples were collected during the period January 2004 to December 2008. CF was diagnosed on the basis of standard methods (sweat chloride test above 60 mmol/l by pilocarpine and two relevant CF transmembrane regulator mutations).
For each patient, data about anthropometric parameters, pancreatic status, and the mean number of pulmonary exacerbations requiring intravenous antibiotics during the previous 12 months were obtained from our existing patient database.
Patients over 6 years of age had at least one lung function evaluation during each year of observation, as measured by the forced expiratory volume in 1 s (FEV1), expressed as a percentage of predicted values for the relevant age, sex, ethnic background, weight, and height.
Despite the central role that pulmonary exacerbations play in CF patient care and research, no standardized definition about pulmonary exacerbations exists. Some of the characteristics most strongly associated with a pulmonary exacerbation include increased cough, increased sputum production, decreased exercise tolerance, decline in weight-for-age percentile, reduced appetite, hemoptysis, and new sounds on examination of the chest [29, 30]. In our patients, pulmonary exacerbations were defined according to these suggestions.
Sputum samples for microbiological studies were obtained from each patient at least every 3 months at clinical examinations. Patients infected by A. xylosoxidans were characterized for age, age of acquisition of first infection, co-infection, lung function, and death. Chronic infection was defined as persistence of three positive cultures for at least six consecutive months, intermittent infection was defined as the presence of three non-consecutive positive cultures per year, and sporadic infection when less than three cultures were positive per year [31]. Co-infection was defined as sputum culture positive for more than one microorganism.
Isolates were defined as multidrug-resistant organisms (MDROs) according to the most recent definition given by the Centers for Disease Control and Prevention (CDC) [32].
In order to correlate chronic A. xylosoxidans infection and chronic P. aeruginosa infection to lung function, clinical features of chronically A. xylosoxidans-infected patients (case group) were compared with those of chronically P. aeruginosa-infected patients (control group); thus, two groups were matched for age, gender, body weight, FEV1, and P. aeruginosa infection status. A. xylosoxidans had never been isolated from any patient of the control group. Nutritional status was calculated as the body mass index (BMI, kg/m2). Data were compared for the period from one year before to one year after the onset of A. xylosoxidans infection. The t-test was used for the analysis of continuous variables and Chi-square test was used for categorical variables. A p-value of less than 0.05 was considered to be statistically significant.
Processing of sputum samples, culture of microorganisms, and phenotypic analysis
Sputum samples, obtained from all patients during the study period, were mixed with equal volumes of 1% dithiothreitol before incubation at 37°C for 30 min. All specimens were examined microscopically and plated on several agar media, including MacConkey agar, CNA agar, BCSA, and Sabouraud agar, at 37°C for 24 up to 72 h.
All isolates were identified by the Phoenix System (Becton Dickinson); the API 20NE identification system (bioMérieux) was used to confirm the identification of A. xylosoxidans isolates.
All A. xylosoxidans isolates were cryopreserved at –80°C for subsequent investigations.
Analysis for 16S rDNA gene-based identification
To validate the phenotypic identification, a polymerase chain reaction (PCR) assay was performed. DNA was prepared by heating one or more colonies (picked from an overnight growth plate) at 95°C for 15 min in 20 μl of lysis buffer containing 0.25% sodium dodecyl sulfate and 0.05 NaOH. After lysis, 180 μl of sterile distilled water was added to the lysis buffer.
The 16S rDNA gene (163 bp) was amplified using the primers AX-F1 and AX-B1, respectively, for the target 5′ and 3′ ends of the 16S rDNA gene locus (AX-F1, sequence 5′-3′ GCAGGAAAGAAACGTCGCGGGT, nucleotide positions 427–448; AX-B1, sequence 5′-3′ ATTTCACATCTTTCTTTCCG, nucleotide positions 576–595) [10]. PCR was performed in a total volume of 100 μl containing 0.5 γ/λ DNA, 25 mM MgCl2, 25 mM dNTPs, 1 μM concentration of each oligonucleotide primer, and 5U/μl Taq polymerase. After initial heating at 94°C for 1 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 45 s, and extension at 72°C for 1 min were performed. The final extension step was at 72°C for 10 min. Control strain (A. xylosoxidans ATCC 27061) and negative control PCRs were employed for every experiment.
Antimicrobial susceptibility test
To assess the sensitivity to aztreonam, piperacillin, piperacillin–tazobactam, cefotaxime, cefepime, ceftazidime, ciprofloxacin, levofloxacin, chloramphenicol, imipenem, meropenem, trimethoprim–sulfamethoxazole, gentamicin, netilmicin, rifampin, and tetracycline, an agar diffusion method (Kirby–Bauer) and the microbroth dilution assay (Phoenix system) were used. A total of five non-CF A. xylosoxidans isolates (one isolate per year of study) was included for the antimicrobial susceptibility analysis. These isolates were obtained from several biological samples, i.e., sputum, blood, vascular catheter, burn wound, and urine.
Interpretative criteria for susceptibility for all of the methods used in the study were in accordance with Clinical and Laboratory Standards Institute (CLSI) criteria [33].
PFGE genotyping
Isolates were grown overnight on nutrient agar and suspended in SE buffer (75 mM NaCl, 25 mM EDTA, pH 7.5). The cell suspension (4 McFarland) was mixed with an equal volume of 1.6% low-melting-point agarose, molded into plugs at 4°C, and lysed with lysis buffer (1% N-lauryl sarcosine, EDTA 0.5 M, pH 8.00), with the addition of proteinase K [34]; the DNA inserts were digested with DraI (Promega), at 37°C for 19 h. Macrorestriction fragments were separated using CHEF DR III (Biorad) at 10°C for 20 h, with start time of 5 s and end-pulse time of 35 s, at a field strength of 6 V/cm. A concatemer ladder of lambda phage DNA was used as a size marker. Fragment patterns were compared according to Tenover’s criteria [35]. Besides, to interpret the molecular patterns, a dendrogram was generated using Phoretix 1D Pro software (TotalLab). To examine whether each patient persistently carried the same A. xylosoxidans strain during the study period, from two up to six isolates for each patient per year were analyzed. A total of five non-CF A. xylosoxidans isolates (one isolate per year of study) was included for the pulsed-field gel electrophoresis (PFGE) analysis (obtained from sputum, blood, vascular catheter, burn wound, and urine).
Results
Prevalence of isolates, chronically infected patients, and co-infection with other microorganisms
Over the study period, a total of 5,315 sputum samples were obtained from 300 patients. In Table 1, the total number of isolates and number and percentage of the total MDROs for each type of bacteria isolated in the study period are reported.
A total of 276 isolates of A. xylosoxidans were recovered and the results obtained by PCR assay were in agreement with biochemical identification. A total of 53 patients (17.6%) had at least two positive cultures for A. xylosoxidans (24 males and 29 females, mean age 21.14 years, range 6.25–48 years).
Among these, 6/53 (11.3%) were defined as chronically infected patients (3 males and 3 females, mean age 11.5 years, range 9.9–21.4 years, mean colonization period 2.7 years) and were co-colonized by P. aeruginosa (mean age at P. aeruginosa chronic infection 9.5 years, range 6–13.7 years).
There were 7/53 (13.2%) patients considered with intermittent infection by A. xylosoxidans (4 males and 3 females, mean age 30 years, range 21–51.9 years), while there were 40/53 (75.5%) patients with sporadic infection (18 males and 22 females, mean age 17.3 years, range 6.2–35.7 years).
In general, co-infection by P. aeruginosa was found in 17/53 patients (32%) with sporadic/intermittent infection. Particularly, 4/17 patients were co-infected by A. xylosoxidans and P. aeruginosa, while 13/17 patients were co-infected by A. xylosoxidans, P. aeruginosa, and other microorganisms (S. aureus, H. influenzae, B. cepacia complex, S. maltophilia, Proteus mirabilis, C. indologenes, and A. fumigatus). Co-infection by Candida albicans was found in 23/53 patients (43.3%).
The correlation among clinical features of chronically A. xylosoxidans-infected patients (case group) and chronically P. aeruginosa-infected patients (control group) was carried out. As shown in Table 2, no significant difference was found between case and control groups in relation to the parameters considered.
Antimicrobial susceptibility test
Considering one isolate per patient, and considering that each isolate showed the same antimicrobial profile during the whole study period, 10/53 (18.8%) A. xylosoxidans isolates were multidrug-resistant, showing resistance to aztreonam (minimum inhibitory concentration MIC > 16 μg/mL), to cephalosporins, including cefepime (MIC > 16 μg/mL), ceftazidime (MIC > 16 μg/mL), and cefotaxime (MIC > 32 μg/mL), to carbapenem (imipenem MIC > 8 μg/mL; meropenem MIC > 8 μg/mL), to aminoglycosides (gentamicin MIC > 8 μg/mL), to ciprofloxacin (MIC > 2 μg/mL), to levofloxacin (MIC > 4 μg/mL), and trimethoprim–sulfamethoxazole (MIC > 2/38 μg/mL). On the other hand, these ten isolates were sensitive to piperacillin (MIC < 4 μg/mL) and piperacillin–tazobactam (MIC < 4/4 μg/mL). All six chronically infected patients carried multidrug-resistant isolates.
Forty-three patients carried isolates that were resistant to aztreonam and sensitive to ceftazidime (MIC < 8 μg/mL), to carbapenem (imipenem MIC < 2 μg/mL; meropenem MIC < 1 μg/mL), to ciprofloxacin (MIC < 2 μg/mL), to levofloxacin (MIC < 24 μg/mL), trimethoprim–sulfamethoxazole (MIC < 0.5/9.5 μg/mL), and chloramphenicol (MIC < 16 μg/mL). Also, these isolates were sensitive to piperacillin and piperacillin–tazobactam. Table 3 shows the results of the antimicrobial susceptibility testing of A. xylosoxidans isolates (n = 53), besides the antimicrobial profile of P. aeruginosa co-infected isolates (n = 23).
Genome macrorestriction analysis
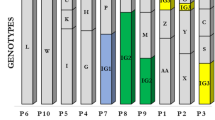
PFGE analysis (Figs. 1 and 2) showed the possibility to group into clusters more than half of the strains (39/53; 73.58%). Seven major clusters were found (A–G clusters). Strains grouped in each cluster showed a diversity (distance level) of less than 10% (or homology level >90%). Cluster A groups 9/53 strains, cluster B groups 3/53 strains, cluster C groups 8/53 strains, cluster D groups 3/53 strains, cluster E groups 4/53 strains, cluster F groups 6/53 strains, and cluster G groups 6/53 strains. The other 14 strains showed a unique macrorestriction profile. Sequential strains obtained in the study period from the same patient showed identical macrorestriction profiles.
Discussion
The survival of CF patients is now increased (median age >30 years) and, consequently, new pathogens have emerged in the CF lung. As underlined by Tan et al. [9], the intensive use of antibiotics may be one factor that increases the likelihood of opportunistic infections with resistant microorganisms.
We found in our cohort a high prevalence of A. xylosoxidans infection (17.6% of the 300 enrolled patients), if the data are compared with those of Magni et al. [36], where a prevalence infection of 8.8% is indicated (enrolled patients = 450). Our data indicate a high prevalence of infection also if they are compared with European studies, such as that conducted by Kanellopoulou et al. [27], where the authors show a prevalence of A. xylosoxidans infection of 12.6% (enrolled patients = 71). The reason for this high prevalence in our population is likely linked to a higher mean age of patients.
The clinical impact of A. xylosoxidans infection is not clear, as well as its lung colonization. Our data are not indicative of increased morbidity linked to this infection/colonization. Besides, our study design does not indicate effects on the clinical status from chronic/intermittent/sporadic A. xylosoxidans infection. We found no significant difference in the FEV1 and BMI comparing chronically A. xylosoxidans infected patients with chronically P. aeruginosa infected patients during the study period. The mild lower mean FEV1 observed in patients with chronic A. xylosoxidans infection could also have been influenced by other covariates, such as diabetes.
All chronically A. xylosoxidans infected patients were co-colonized also by P. aeruginosa and, generally, 43.3% of patients present also P. aeruginosa infection. In the study of Van Daele et al. [37], there is indicated the strong tendency by A. xylosoxidans to install itself in a lung already infected by P. aeruginosa. But, in this study, only patients colonized by P. aeruginosa were enrolled. Our data also indicate this tendency by A. xylosoxidans but, differently from the study of Van Daele et al., our population also included patients not co-colonized by P. aeruginosa (56.6%; 30 non-co-infected by P. aeruginosa/53). Thus, we can affirm that A. xylosoxidans can infect a lung also not previously colonized by P. aeruginosa.
In our cohort, none of the patients had a transplantation during the study. About 10% of patients (5/53) died in the examined period, two were chronically infected by A. xylosoxidans and all of these five patients were chronically infected by P. aeruginosa. However, it is not possible to link these deaths to A. xylosoxidans infection/colonization.
Because of the emergence of new Gram-negative microorganisms among CF patients, bacterial misidentification is especially problematic and presents a challenge to effective infection control in CF, to antimicrobial therapy, and to patient prognosis.
There is evidence of weakness of commercial systems for the phenotypic identification of non-fermentative Gram-negative bacteria recovered from CF patients. Saiman et al., in a report from 2001 [26], indicated that, in a group of 106 isolates obtained from 78 patients from 49 CF centers in the United States and all identified as A. xylosoxidans, 89% were correctly identified, whereas 11% were misidentified; thus, ten isolates were found to be P. aeruginosa, one isolate S. maltophilia, and one B. cepacia complex. We have demonstrated that the phenotypic identification, carried out by Phoenix and API 20NE systems, was confirmed by PCR analysis.
As a consequence of the increasing use of antibiotics concomitant to acute pulmonary exacerbations in CF patients due to P. aeruginosa infection, A. xylosoxidans as well as other non-fermentative Gram-negative bacteria are showing growing drug resistance. Our data support this evidence because of frequent previous colonization with P. aeruginosa.
It is well known that, for most of the non-lactose fermenting, Gram-negative rods, the disk diffusion antibiogram is not validated by the CLSI. In fact, there are several interpretation problems, such as unclear inhibition zone borders. Consequently, in the present study, a microbroth dilution assay was also carried out and no differences were found between the two methods.
With the emergence of antimicrobial resistance of new pathogens, the scenario has changed with respect to the availability and susceptibility of antimicrobial agents. In our study, about 20% of A. xylosoxidans isolates showed a multidrug-resistant profile. This percentage of isolates was resistant to aztreonam, cephalosporins, including cefepime, ceftazidime, and cefotaxime, to carbapenem, aminoglycosides, quinolones, and trimethoprim–sulfamethoxazole. These same isolates were sensitive to piperacillin with or without tazobactam. Ceftazidime, carbapenems, quinolones, and trimethoprim–sulfamethoxazole, besides piperacillin and piperacillin–tazobactam, were active against non-multidrug-resistant isolates. A. xylosoxidans isolates recovered from non-CF patients were resistant to molecules such as ceftazidime, carbapenems, and levofloxacin. This finding indicates that isolates of A. xylosoxidans can be resistant to many antimicrobial agents, independently from the study population.
Currently, reports for patient-to-patient spread are controversial. Several studies rule out the possibility of A. xylosoxidans by inter-human contacts, or at least they indicate little evidence of person-to-person transmission, such as the study of Vu-Thien et al. [22]. In our study, PFGE indicates that more than half of A. xylosoxidans can be grouped into seven different clusters, suggesting the possibility of patient-to-patient transmission.
The results of PFGE have been interesting for several reasons. First, all six chronically infected patients carried strains of two different clusters (four strains of cluster A and two strains of cluster C); besides, among five cases of death, one patient carried strain A and four patients carried strains with a unique macro-restriction profile. At last, any strain with a unique macro-restriction profile was considered to be an MDRO, whereas four strains of clone A, two of clone B, and four of clone C were considered to be MDROs.
These results are supported by a recent study of Kanellopoulou et al. [27], where the authors indicated five CF patients colonized by genetically related strains. Our results certainly suggest a common source of contamination and this is highly probable because of the frequent social contacts that occur among patients of a single care center.
It is very interesting to note that, in our study, restriction patterns were obtained by both DraI and by SmaI. Restriction endonuclease SmaI generated a lower number of bands with respect to those generated by DraI (data not shown). Consequently, DNA cleavage by this endonuclease was characterized by increased sensitivity.
In conclusion, the results of the present study can represent a further step toward the understanding of the epidemiology of these microorganisms and of a possible correlation between the microbiological data and clinical outcomes of CF patients.
References
Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, Ramsey BW, Clausen CR (1998) Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis 27(1):158–163
Lyczak JB, Cannon CL, Pier GB (2002) Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15(2):194–222
Rajan S, Saiman L (2002) Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect 17(1):47–56
Saiman L, Siegel J (2004) Infection control in cystic fibrosis. Clin Microbiol Rev 17(1):57–71
Lambiase A, Raia V, Del Pezzo M, Sepe A, Carnovale V, Rossano F (2006) Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 6:4
Frederiksen B, Koch C, Høiby N (1999) Changing epidemiology of Pseudomonas aeruginosa infection in Danish cystic fibrosis patients (1974–1995). Pediatr Pulmonol 28(3):159–166
Lambiase A, Raia V, Stefani S, Sepe A, Ferri P, Buonpensiero P, Rossano F, Del Pezzo M (2007) Burkholderia cepacia complex infection in a cohort of Italian patients with cystic fibrosis. J Microbiol 45(3):275–279
Goss CH, Otto K, Aitken ML, Rubenfeld GD (2002) Detecting Stenotrophomonas maltophilia does not reduce survival of patients with cystic fibrosis. Am J Respir Crit Care Med 166(3):356–361
Tan K, Conway SP, Brownlee KG, Etherington C, Peckham DG (2002) Alcaligenes infection in cystic fibrosis. Pediatr Pulmonol 34:101–104
Liu L, Coenye T, Burns JL, Whitby PW, Stull TL, LiPuma JJ (2002) Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J Clin Microbiol 40:1210–1213
Bakare N, Rickerts V, Bargon J, Just-Nübling G (2003) Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 46(1–2):19–23
Olivier KN, Weber DJ, Lee JH, Handler A, Tudor G, Molina PL, Tomashefski J, Knowles MR; Nontuberculous Mycobacteria in Cystic Fibrosis Study Group (2003) Nontuberculous mycobacteria. II: nested-cohort study of impact on cystic fibrosis lung disease. Am J Respir Crit Care Med 167(6):835–840
Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ (2002) Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol 40(6):2062–2069
Lambiase A, Del Pezzo M, Raia V, Sepe A, Ferri P, Rossano F (2007) Chryseobacterium respiratory tract infections in patients with cystic fibrosis. J Infect 55(6):518–23
Lambiase A, Rossano F, Del Pezzo M, Raia V, Sepe A, de Gregorio F, Catania MR (2009) Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res Notes 2:262
Cimon B, Carrère J, Vinatier JF, Chazalette JP, Chabasse D, Bouchara JP (2000) Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 19(1):53–56
Defontaine A, Zouhair R, Cimon B, Carrère J, Bailly E, Symoens F, Diouri M, Hallet JN, Bouchara JP (2002) Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J Clin Microbiol 40(6):2108–2114
Cimon B, Carrère J, Chazalette JP, Vinatier JF, Chabasse D, Bouchara JP (1999) Chronic airway colonization by Penicillium emersonii in a patient with cystic fibrosis. Med Mycol 37(4):291–293
Pihet M, Carrère J, Cimon B, Chabasse D, Delhaes L, Symoens F, Bouchara JP (2009) Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—a review. Med Mycol 47(4):387–397
Yabuuchi E, Kawamura Y, Kosako Y, Ezaki T (1998) Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Rüger and Tan) comb. nov. Microbiol Immunol 42:429–438
Manfredi R, Nanetti A, Ferri M, Chiodo F (1997) Bacteremia and respiratory involvement by Alcaligenes xylosoxidans in patients infected with the human immunodeficiency virus. Eur J Clin Microbiol Infect Dis 16:933–938
Vu-Thien H, Darbord JC, Moissenet D, Dulot C, Dufourcq JB, Marsol P, Garbarg-Chenon A (1998) Investigation of an outbreak of wound infections due to Alcaligenes xylosoxidans transmitted by chlorhexidine in a burns unit. Eur J Clin Microbiol Infect Dis 17:724–726
Tena D, Carranza R, Barberá JR, Valdezate S, Garrancho JM, Arranz M, Sáez-Nieto JA (2005) Outbreak of long-term intravascular catheter-related bacteremia due to Achromobacter xylosoxidans subspecies xylosoxidans in a hemodialysis unit. Eur J Clin Microbiol Infect Dis 24:727–732
Peltroche-Llacshuanga H, Kentrup H, Haase G (1998) Persistent airway colonization with Alcaligenes xylosoxidans in two brothers with cystic fibrosis. Eur J Clin Microbiol Infect Dis 17:132–134
Vu-Thien H, Moissenet D, Valcin M, Dulot C, Tournier G, Garbarg-Chenon A (1996) Molecular epidemiology of Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans in a cystic fibrosis center. Eur J Clin Microbiol Infect Dis 15:876–879
Saiman L, Chen Y, Tabibi S, San Gabriel P, Zhou J, Liu Z, Lai L, Whittier S (2001) Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J Clin Microbiol 39:3942–3945
Kanellopoulou M, Pournaras S, Iglezos H, Skarmoutsou N, Papafrangas E, Maniatis AN (2004) Persistent colonization of nine cystic fibrosis patients with an Achromobacter (Alcaligenes) xylosoxidans clone. Eur J Clin Microbiol Infect Dis 23:336–339
Raso T, Bianco O, Grosso B, Zucca M, Savoia D (2008) Achromobacter xylosoxidans respiratory tract infections in cystic fibrosis patients. APMIS 116:837–841
Ramsey BW (1996) Management of pulmonary disease in patients with cystic fibrosis. N Engl J Med 335:179–188
Saiman L, MacDonald NE, Whittier S (1996) The microbiological basis for antimicrobial therapy. In: Proceedings of the 10th Annual North American Cystic Fibrosis Conference, Orlando, FL, October 1996. Cystic Fibrosis Foundation
Frederiksen B, Koch C, Høiby N (1999) Changing epidemiology of Pseudomonas aeruginosa infection in Danish cystic fibrosis patients (1974–1995). Pediatr Pulmonol 28:159–166
Siegel JD, Rhinehart E, Jackson M, Chiarello L; The Healthcare Infection Control Practices Advisory Committee (2006) Management of multidrug-resistant organisms in healthcare settings, 2006. Centers for Disease Control and Prevention (CDC), pp 1–74. Available online at: http://www.cdc.gov/ncidod/dhqp/pdf/ar/MDROGuideline2006.pdf
National Committee for Clinical Laboratory Standards (NCCLS) (2002) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th edn. Approved Standard M7-A5. NCCLS, Wayne, PA
Grothues D, Koopmann U, von der Hardt H, Tümmler B (1988) Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol 26:1973–1977
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239
Magni A, Trancassini M, Varesi P, Iebba V, Curci A, Pecoraro C, Cimino G, Schippa S, Quattrucci S (2010) Achromobacter xylosoxidans genomic characterization and correlation of randomly amplified polymorphic DNA profiles with relevant clinical features [corrected] of cystic fibrosis patients. J Clin Microbiol 48:1035–1039
Van Daele S, Verhelst R, Claeys G, Verschraegen G, Franckx H, Van Simaey L, de Ganck C, De Baets F, Vaneechoutte M (2005) Shared genotypes of Achromobacter xylosoxidans strains isolated from patients at a cystic fibrosis rehabilitation center. J Clin Microbiol 43:2998–3002
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lambiase, A., Catania, M.R., del Pezzo, M. et al. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis 30, 973–980 (2011). https://doi.org/10.1007/s10096-011-1182-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-011-1182-5